
| Name: LSM4 | Sequence: fasta or formatted (139aa) | NCBI GI: 6912486 | |
|
Description: U6 snRNA-associated Sm-like protein 4
|
Referenced in: RNases and RNA Stability
| ||
|
Composition:
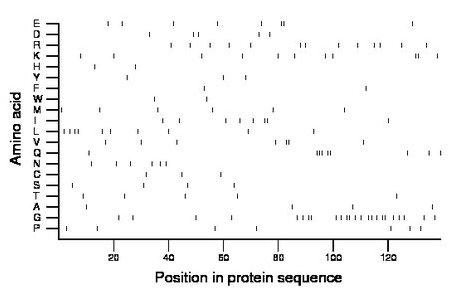
Amino acid Percentage Count Longest homopolymer A alanine 2.9 4 1 C cysteine 2.2 3 1 D aspartate 3.6 5 1 E glutamate 5.8 8 2 F phenylalanine 1.4 2 1 G glycine 17.3 24 2 H histidine 1.4 2 1 I isoleucine 5.8 8 2 K lysine 7.9 11 2 L leucine 7.2 10 2 M methionine 4.3 6 1 N asparagine 4.3 6 1 P proline 5.0 7 1 Q glutamine 6.5 9 3 R arginine 9.4 13 1 S serine 2.9 4 1 T threonine 3.6 5 1 V valine 5.0 7 2 W tryptophan 1.4 2 1 Y tyrosine 2.2 3 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 U6 snRNA-associated Sm-like protein 4 SNRPD3 0.142 small nuclear ribonucleoprotein polypeptide D3 SNRPD1 0.104 small nuclear ribonucleoprotein D1 polypeptide 16kDa ... LSM2 0.086 LSM2 homolog, U6 small nuclear RNA associated LOC100129492 0.071 PREDICTED: similar to small nuclear ribonucleoprote... C1orf77 0.067 small protein rich in arginine and glycine NCL 0.063 nucleolin LSM10 0.063 LSM10, U7 small nuclear RNA associated BRWD3 0.060 bromodomain and WD repeat domain containing 3 [Homo... MBD2 0.056 methyl-CpG binding domain protein 2 testis-specific i... MBD2 0.056 methyl-CpG binding domain protein 2 isoform 1 AVEN 0.056 cell death regulator aven FBL 0.052 fibrillarin FUS 0.049 fusion (involved in t(12;16) in malignant liposarcoma... SNRPF 0.049 small nuclear ribonucleoprotein polypeptide F MESP2 0.049 mesoderm posterior 2 homolog FAM98B 0.045 family with sequence similarity 98, member B isofor... HNRNPA1 0.037 heterogeneous nuclear ribonucleoprotein A1 isoform b... HNRNPA1 0.037 heterogeneous nuclear ribonucleoprotein A1 isoform a ... GAR1 0.037 nucleolar protein family A, member 1 GAR1 0.037 nucleolar protein family A, member 1 ZC3H4 0.037 zinc finger CCCH-type containing 4 LSM6 0.034 Sm protein F HNRPDL 0.034 heterogeneous nuclear ribonucleoprotein D-like [Homo... RRS1 0.030 homolog of yeast ribosome biogenesis regulatory prot... XYLT1 0.030 xylosyltransferase I LOC100291596 0.030 PREDICTED: hypothetical protein XP_002344746 LOC100288068 0.030 PREDICTED: hypothetical protein LOC100288068 0.030 PREDICTED: hypothetical protein XP_002343259 LOC100292082 0.026 PREDICTED: hypothetical protein XP_002345881Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
