
| Name: PDAP1 | Sequence: fasta or formatted (181aa) | NCBI GI: 7657441 | |
|
Description: PDGFA associated protein 1
|
Referenced in:
| ||
|
Composition:
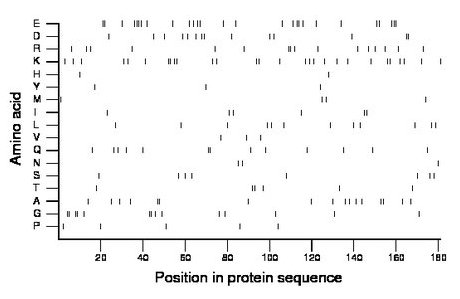
Amino acid Percentage Count Longest homopolymer A alanine 9.4 17 2 C cysteine 0.0 0 0 D aspartate 7.7 14 2 E glutamate 14.9 27 4 F phenylalanine 0.0 0 0 G glycine 7.7 14 2 H histidine 1.1 2 1 I isoleucine 3.3 6 2 K lysine 15.5 28 2 L leucine 6.6 12 1 M methionine 2.2 4 1 N asparagine 1.7 3 1 P proline 2.8 5 1 Q glutamine 7.2 13 2 R arginine 8.8 16 2 S serine 4.4 8 1 T threonine 3.3 6 2 V valine 1.7 3 1 W tryptophan 0.0 0 0 Y tyrosine 1.7 3 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PDGFA associated protein 1 KIF3B 0.088 kinesin family member 3B TRDN 0.088 triadin LOC100131202 0.085 PREDICTED: hypothetical protein LOC100133599 0.085 PREDICTED: hypothetical protein RDX 0.076 radixin LOC100131202 0.073 PREDICTED: hypothetical protein TCHH 0.073 trichohyalin CYLC2 0.070 cylicin 2 LOC100286959 0.070 PREDICTED: hypothetical protein XP_002343921 PES1 0.070 pescadillo homolog 1, containing BRCT domain RBBP6 0.067 retinoblastoma-binding protein 6 isoform 2 RBBP6 0.067 retinoblastoma-binding protein 6 isoform 1 ASPH 0.067 aspartate beta-hydroxylase isoform d ASPH 0.067 aspartate beta-hydroxylase isoform e KIAA1211 0.064 hypothetical protein LOC57482 MSN 0.061 moesin EIF5B 0.061 eukaryotic translation initiation factor 5B TNNT2 0.061 troponin T type 2, cardiac isoform 4 TNNT2 0.061 troponin T type 2, cardiac isoform 3 TNNT2 0.061 troponin T type 2, cardiac isoform 2 TNNT2 0.061 troponin T type 2, cardiac isoform 1 DDX23 0.061 DEAD (Asp-Glu-Ala-Asp) box polypeptide 23 LOC100133599 0.058 PREDICTED: hypothetical protein LOC100288837 0.058 PREDICTED: hypothetical protein XP_002343931, parti... PTMA 0.058 prothymosin, alpha isoform 2 YTHDC1 0.058 splicing factor YT521-B isoform 2 YTHDC1 0.058 splicing factor YT521-B isoform 1 LOC100294088 0.058 PREDICTED: hypothetical protein, partial NUMA1 0.058 nuclear mitotic apparatus protein 1Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
