
| Name: RIPPLY2 | Sequence: fasta or formatted (128aa) | NCBI GI: 58000465 | |
|
Description: ripply2 protein
|
Referenced in:
| ||
|
Composition:
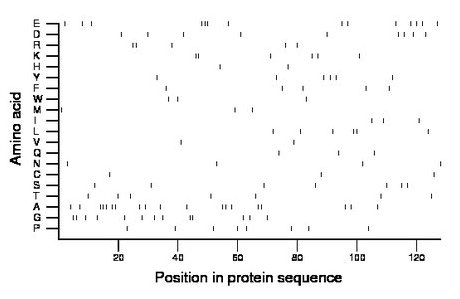
Amino acid Percentage Count Longest homopolymer A alanine 14.8 19 3 C cysteine 2.3 3 1 D aspartate 7.0 9 1 E glutamate 10.9 14 3 F phenylalanine 3.9 5 1 G glycine 10.2 13 2 H histidine 1.6 2 1 I isoleucine 2.3 3 1 K lysine 4.7 6 2 L leucine 4.7 6 2 M methionine 2.3 3 1 N asparagine 3.1 4 1 P proline 6.2 8 1 Q glutamine 2.3 3 1 R arginine 3.9 5 2 S serine 5.5 7 1 T threonine 5.5 7 1 V valine 1.6 2 1 W tryptophan 2.3 3 1 Y tyrosine 4.7 6 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 ripply2 protein RIPPLY1 0.246 ripply1 homolog DSCR6 0.161 Down syndrome critical region protein 6 FBF1 0.036 Fas (TNFRSF6) binding factor 1 NUP62 0.020 nucleoporin 62kDa NUP62 0.020 nucleoporin 62kDa NUP62 0.020 nucleoporin 62kDa NUP62 0.020 nucleoporin 62kDa FAM190B 0.020 granule cell antiserum positive 14 EVPLL 0.016 envoplakin-like C19orf22 0.016 hypothetical protein LOC91300 LOC100287063 0.016 PREDICTED: hypothetical protein YLPM1 0.016 YLP motif containing 1 HNRNPD 0.012 heterogeneous nuclear ribonucleoprotein D isoform b ... HNRNPD 0.012 heterogeneous nuclear ribonucleoprotein D isoform d ... DDX51 0.012 DEAD (Asp-Glu-Ala-Asp) box polypeptide 51 LOC100289898 0.012 PREDICTED: hypothetical protein XP_002346876 LOC100289162 0.012 PREDICTED: hypothetical protein XP_002342718 HOXA11 0.012 homeobox A11 KCNH1 0.012 potassium voltage-gated channel, subfamily H, member ... KCNH1 0.012 potassium voltage-gated channel, subfamily H, member... HNRNPD 0.012 heterogeneous nuclear ribonucleoprotein D isoform a ... HNRNPD 0.012 heterogeneous nuclear ribonucleoprotein D isoform c ... LMTK3 0.012 lemur tyrosine kinase 3 C9orf75 0.012 hypothetical protein LOC286262 isoform 2 C9orf75 0.012 hypothetical protein LOC286262 isoform 1 NAP1L5 0.008 nucleosome assembly protein 1-like 5 CHD1 0.008 chromodomain helicase DNA binding protein 1 LOC100290709 0.008 PREDICTED: hypothetical protein XP_002346752 TLX2 0.008 T-cell leukemia homeobox 2Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
