
| Name: LOC100289898 | Sequence: fasta or formatted (552aa) | NCBI GI: 239748993 | |
|
Description: PREDICTED: hypothetical protein XP_002346876
| Not currently referenced in the text | ||
|
Composition:
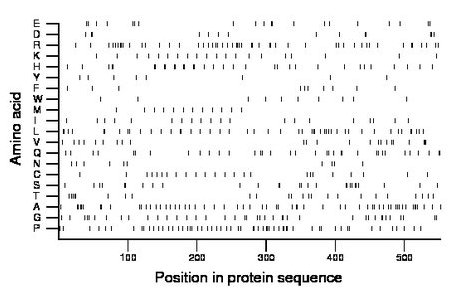
Amino acid Percentage Count Longest homopolymer A alanine 10.7 59 2 C cysteine 4.3 24 1 D aspartate 2.5 14 2 E glutamate 3.6 20 2 F phenylalanine 1.8 10 1 G glycine 7.2 40 2 H histidine 6.3 35 2 I isoleucine 3.6 20 1 K lysine 3.6 20 1 L leucine 9.1 50 2 M methionine 2.4 13 1 N asparagine 1.8 10 1 P proline 10.5 58 2 Q glutamine 6.3 35 2 R arginine 9.2 51 2 S serine 4.9 27 2 T threonine 4.3 24 1 V valine 4.2 23 3 W tryptophan 2.0 11 2 Y tyrosine 1.4 8 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002346876 LOC100289162 0.995 PREDICTED: hypothetical protein XP_002342718 LOC100293694 0.412 PREDICTED: hypothetical protein LOC100287903 0.014 PREDICTED: hypothetical protein XP_002343649 LOC100134663 0.011 PREDICTED: hypothetical protein, partial NDN 0.010 necdin LOC100287936 0.010 PREDICTED: hypothetical protein XP_002343650 LOC643355 0.010 PREDICTED: hypothetical protein RERE 0.010 atrophin-1 like protein isoform b RERE 0.010 atrophin-1 like protein isoform a RERE 0.010 atrophin-1 like protein isoform a CNGB1 0.009 cyclic nucleotide gated channel beta 1 isoform b [H... CNGB1 0.009 cyclic nucleotide gated channel beta 1 isoform a [H... SATL1 0.009 spermidine/spermine N1-acetyl transferase-like 1 [Ho... LOC100287484 0.009 PREDICTED: hypothetical protein XP_002343105 ZNF469 0.008 zinc finger protein 469 ZMIZ1 0.007 retinoic acid induced 17 LOC100133500 0.007 PREDICTED: hypothetical protein LOC100293662 0.007 PREDICTED: hypothetical protein LOC100289444 0.007 PREDICTED: hypothetical protein LOC100289444 0.007 PREDICTED: hypothetical protein XP_002342754 LOC100289444 0.007 PREDICTED: hypothetical protein NEFH 0.007 neurofilament, heavy polypeptide 200kDa LRRC56 0.007 leucine rich repeat containing 56 LOC388906 0.007 PREDICTED: hypothetical protein LOC388906 0.007 PREDICTED: hypothetical protein LOC388906 0.007 PREDICTED: hypothetical protein HGC6.3 0.007 hypothetical protein LOC100128124 LOC100291911 0.006 PREDICTED: similar to Putative uncharacterized prot... CREB5 0.006 cAMP responsive element binding protein 5 isoform de...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
