
| Name: MED26 | Sequence: fasta or formatted (600aa) | NCBI GI: 28558977 | |
|
Description: mediator complex subunit 26
|
Referenced in: Nuclear Receptors
| ||
|
Composition:
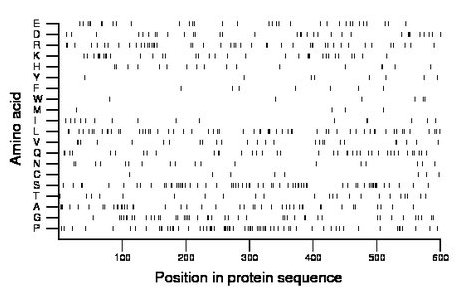
Amino acid Percentage Count Longest homopolymer A alanine 7.3 44 2 C cysteine 1.3 8 1 D aspartate 5.7 34 2 E glutamate 5.7 34 2 F phenylalanine 1.2 7 1 G glycine 7.3 44 2 H histidine 2.8 17 2 I isoleucine 2.7 16 2 K lysine 5.8 35 4 L leucine 9.8 59 2 M methionine 0.8 5 1 N asparagine 3.0 18 1 P proline 11.0 66 3 Q glutamine 6.7 40 2 R arginine 7.7 46 2 S serine 10.7 64 3 T threonine 3.8 23 1 V valine 4.3 26 1 W tryptophan 1.0 6 1 Y tyrosine 1.3 8 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 mediator complex subunit 26 COL1A1 0.028 alpha 1 type I collagen preproprotein TCEA1 0.028 transcription elongation factor A 1 isoform 1 LOC100289899 0.026 PREDICTED: hypothetical protein XP_002347041 TCEA2 0.023 transcription elongation factor A protein 2 isoform a... COL1A2 0.023 alpha 2 type I collagen PRB1 0.023 proline-rich protein BstNI subfamily 1 isoform 1 pre... COL3A1 0.022 collagen type III alpha 1 preproprotein SRRM1 0.022 serine/arginine repetitive matrix 1 TCEA3 0.021 transcription elongation factor A (SII), 3 TCEA2 0.021 transcription elongation factor A protein 2 isoform ... SYNJ1 0.020 synaptojanin 1 isoform d ZC3H4 0.020 zinc finger CCCH-type containing 4 MAP7D1 0.020 MAP7 domain containing 1 MLL2 0.019 myeloid/lymphoid or mixed-lineage leukemia 2 BAT2 0.019 HLA-B associated transcript-2 COL4A4 0.019 alpha 4 type IV collagen precursor COL19A1 0.019 alpha 1 type XIX collagen precursor KIAA1522 0.018 hypothetical protein LOC57648 COL22A1 0.018 collagen, type XXII, alpha 1 SYDE1 0.018 synapse defective 1, Rho GTPase, homolog 1 KIF26B 0.018 kinesin family member 26B COL8A2 0.018 collagen, type VIII, alpha 2 PRB2 0.018 proline-rich protein BstNI subfamily 2 COL23A1 0.018 collagen, type XXIII, alpha 1 COL7A1 0.018 alpha 1 type VII collagen precursor COL2A1 0.018 collagen, type II, alpha 1 isoform 1 precursor [Hom... COL2A1 0.018 collagen, type II, alpha 1 isoform 2 precursor [Hom... LOC100131102 0.018 PREDICTED: hypothetical protein COL9A1 0.018 alpha 1 type IX collagen isoform 1 precursorHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
