
| Name: LOC100288835 | Sequence: fasta or formatted (248aa) | NCBI GI: 239742745 | |
|
Description: PREDICTED: hypothetical protein XP_002342616
| Not currently referenced in the text | ||
|
Composition:
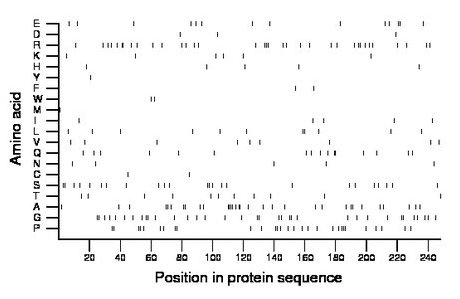
Amino acid Percentage Count Longest homopolymer A alanine 12.9 32 4 C cysteine 0.8 2 1 D aspartate 1.2 3 1 E glutamate 5.6 14 2 F phenylalanine 0.8 2 1 G glycine 14.1 35 2 H histidine 2.0 5 1 I isoleucine 2.0 5 1 K lysine 2.4 6 1 L leucine 4.4 11 2 M methionine 0.4 1 1 N asparagine 2.0 5 1 P proline 11.7 29 3 Q glutamine 6.5 16 2 R arginine 14.1 35 3 S serine 9.7 24 2 T threonine 4.4 11 1 V valine 3.6 9 1 W tryptophan 0.8 2 1 Y tyrosine 0.4 1 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002342616 LOC100290139 0.843 PREDICTED: hypothetical protein XP_002346782 PRB1 0.061 proline-rich protein BstNI subfamily 1 isoform 1 pre... PRB4 0.054 proline-rich protein BstNI subfamily 4 precursor [Ho... FLJ37078 0.052 hypothetical protein LOC222183 LOC100170229 0.052 hypothetical protein LOC100170229 PRB2 0.050 proline-rich protein BstNI subfamily 2 PRB3 0.050 proline-rich protein BstNI subfamily 3 precursor [H... SAMD1 0.048 sterile alpha motif domain containing 1 ZNF469 0.046 zinc finger protein 469 COL7A1 0.044 alpha 1 type VII collagen precursor BSN 0.044 bassoon protein SYNJ1 0.042 synaptojanin 1 isoform b COL1A1 0.042 alpha 1 type I collagen preproprotein SNRPB 0.042 small nuclear ribonucleoprotein polypeptide B/B' iso... OGFR 0.042 opioid growth factor receptor LOC100293005 0.042 PREDICTED: hypothetical protein LOC440829 0.042 PREDICTED: transmembrane protein 46-like ANKRD33B 0.042 PREDICTED: ankyrin repeat domain 33B FBLN2 0.042 fibulin 2 isoform b precursor FBLN2 0.042 fibulin 2 isoform a precursor TAF4 0.040 TBP-associated factor 4 COL5A1 0.040 alpha 1 type V collagen preproprotein COL3A1 0.040 collagen type III alpha 1 preproprotein LOC100288108 0.040 PREDICTED: hypothetical protein LOC100288108 0.040 PREDICTED: hypothetical protein XP_002342919 SCAF1 0.040 SR-related CTD-associated factor 1 LOC100293375 0.038 PREDICTED: hypothetical protein MAMSTR 0.038 MEF2 activating motif and SAP domain containing tra... MAMSTR 0.038 MEF2 activating motif and SAP domain containing tran...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
