
| Name: LOC100132388 | Sequence: fasta or formatted (107aa) | NCBI GI: 169210998 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
Other entries for this name:
alt prot {107aa} PREDICTED: hypothetical protein alt mRNA {107aa} PREDICTED: hypothetical protein | |||
|
Composition:
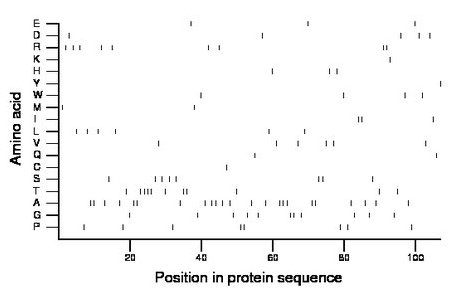
Amino acid Percentage Count Longest homopolymer A alanine 21.5 23 3 C cysteine 0.9 1 1 D aspartate 4.7 5 1 E glutamate 2.8 3 1 F phenylalanine 0.0 0 0 G glycine 10.3 11 2 H histidine 2.8 3 1 I isoleucine 2.8 3 2 K lysine 0.9 1 1 L leucine 5.6 6 1 M methionine 1.9 2 1 N asparagine 0.0 0 0 P proline 7.5 8 2 Q glutamine 1.9 2 1 R arginine 8.4 9 2 S serine 7.5 8 2 T threonine 10.3 11 4 V valine 5.6 6 1 W tryptophan 3.7 4 1 Y tyrosine 0.9 1 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC100132388 1.000 PREDICTED: hypothetical protein LOC100132388 0.990 PREDICTED: hypothetical protein AMOT 0.051 angiomotin isoform 2 AMOT 0.051 angiomotin isoform 1 HCN2 0.031 hyperpolarization activated cyclic nucleotide-gated... GLCCI1 0.026 glucocorticoid induced transcript 1 TP53INP1 0.026 tumor protein p53 inducible nuclear protein 1 isofor... HMX1 0.026 homeo box (H6 family) 1 SOX21 0.026 SRY-box 21 LOC100292407 0.026 PREDICTED: hypothetical protein RICH2 0.026 Rho GTPase-activating protein RICH2 SHISA3 0.021 shisa homolog 3 IGSF9B 0.021 immunoglobulin superfamily, member 9B MLL 0.021 myeloid/lymphoid or mixed-lineage leukemia protein [... LOC100290358 0.021 PREDICTED: hypothetical protein XP_002346903 LOC100294079 0.021 PREDICTED: hypothetical protein MUC5B 0.021 mucin 5, subtype B, tracheobronchial SFRS15 0.021 splicing factor, arginine/serine-rich 15 isoform 3 ... SFRS15 0.021 splicing factor, arginine/serine-rich 15 isoform 2 ... SFRS15 0.021 splicing factor, arginine/serine-rich 15 isoform 1 [... PPHLN1 0.021 periphilin 1 isoform 6 PPHLN1 0.021 periphilin 1 isoform 1 PPHLN1 0.021 periphilin 1 isoform 4 PPHLN1 0.021 periphilin 1 isoform 3 PPHLN1 0.021 periphilin 1 isoform 5 PPHLN1 0.021 periphilin 1 isoform 2 ISCA2 0.021 iron-sulfur cluster assembly 2 HOXD9 0.015 homeobox D9 LOC100132635 0.015 PREDICTED: similar to mucin 5, partialHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
