
| Name: LOC100131583 | Sequence: fasta or formatted (208aa) | NCBI GI: 169177938 | |
|
Description: PREDICTED: similar to chromosome 9 open reading frame 147
| Not currently referenced in the text | ||
|
Composition:
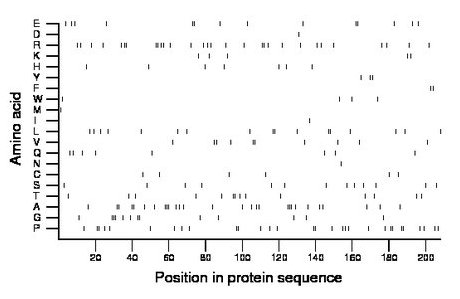
Amino acid Percentage Count Longest homopolymer A alanine 13.5 28 3 C cysteine 2.9 6 1 D aspartate 0.5 1 1 E glutamate 6.7 14 2 F phenylalanine 1.0 2 2 G glycine 6.2 13 3 H histidine 3.4 7 1 I isoleucine 0.5 1 1 K lysine 2.4 5 1 L leucine 8.2 17 2 M methionine 0.5 1 1 N asparagine 0.5 1 1 P proline 14.4 30 2 Q glutamine 3.8 8 1 R arginine 14.4 30 2 S serine 6.2 13 1 T threonine 6.7 14 2 V valine 4.8 10 2 W tryptophan 1.9 4 1 Y tyrosine 1.4 3 2 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: similar to chromosome 9 open reading fra... LOC643355 0.049 PREDICTED: hypothetical protein LOC643355 0.049 PREDICTED: hypothetical protein LOC643355 0.046 PREDICTED: hypothetical protein SRRM1 0.034 serine/arginine repetitive matrix 1 LOC727804 0.034 PREDICTED: hypothetical protein LOC100131102 0.029 PREDICTED: hypothetical protein ZNF469 0.029 zinc finger protein 469 SH3RF1 0.024 SH3 domain containing ring finger 1 ASXL3 0.024 additional sex combs like 3 FLJ22184 0.024 PREDICTED: hypothetical protein LOC80164 HNRPDL 0.024 heterogeneous nuclear ribonucleoprotein D-like [Homo... OTUD7A 0.024 OTU domain containing 7A TTLL8 0.024 tubulin tyrosine ligase-like family, member 8 [Homo... FLJ22184 0.022 PREDICTED: hypothetical protein FLJ22184 LOC100287499 0.022 PREDICTED: hypothetical protein LOC100287499 0.022 PREDICTED: hypothetical protein XP_002343643 FLJ22184 0.022 PREDICTED: hypothetical protein FLJ22184 KCNH3 0.022 potassium voltage-gated channel, subfamily H (eag-re... LOC441617 0.019 PREDICTED: similar to hCG1820409 LOC441617 0.019 PREDICTED: similar to hCG1820409 LOC100292801 0.019 PREDICTED: hypothetical protein LOC100290691 0.019 PREDICTED: hypothetical protein XP_002347934 LOC100131102 0.019 PREDICTED: hypothetical protein LOC100288156 0.019 PREDICTED: hypothetical protein XP_002344202 LOC100131102 0.019 PREDICTED: hypothetical protein LOC644578 0.019 PREDICTED: similar to hCG2040199 ARL6IP4 0.019 SRp25 nuclear protein isoform 3 LOC644578 0.019 PREDICTED: similar to hCG2040199 LOC100130360 0.019 PREDICTED: hypothetical proteinHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
