
| Name: FAM83H | Sequence: fasta or formatted (1179aa) | NCBI GI: 157311635 | |
|
Description: FAM83H
| Not currently referenced in the text | ||
|
Composition:
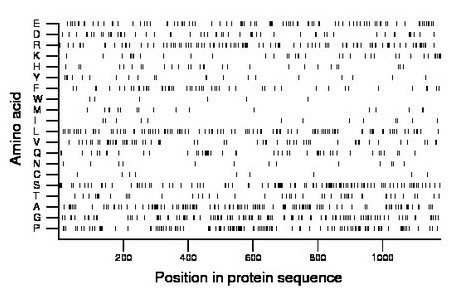
Amino acid Percentage Count Longest homopolymer A alanine 10.5 124 4 C cysteine 0.8 9 1 D aspartate 4.8 57 2 E glutamate 7.8 92 2 F phenylalanine 4.3 51 2 G glycine 9.8 116 3 H histidine 2.5 29 2 I isoleucine 1.1 13 1 K lysine 3.3 39 2 L leucine 8.9 105 2 M methionine 1.5 18 1 N asparagine 1.1 13 1 P proline 10.2 120 4 Q glutamine 4.0 47 3 R arginine 8.7 103 2 S serine 9.2 108 3 T threonine 3.8 45 2 V valine 5.2 61 2 W tryptophan 0.5 6 1 Y tyrosine 2.0 23 2 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 FAM83H FAM83B 0.099 hypothetical protein LOC222584 FAM83C 0.069 hypothetical protein LOC128876 FAM83E 0.068 hypothetical protein LOC54854 FAM83G 0.065 hypothetical protein LOC644815 FAM83D 0.063 hypothetical protein LOC81610 FAM83A 0.062 hypothetical protein LOC84985 isoform a FAM83F 0.060 hypothetical protein LOC113828 FAM83A 0.060 hypothetical protein LOC84985 isoform b SPEG 0.014 SPEG complex locus APC2 0.012 adenomatosis polyposis coli 2 COL2A1 0.011 collagen, type II, alpha 1 isoform 1 precursor [Hom... COL2A1 0.011 collagen, type II, alpha 1 isoform 2 precursor [Hom... COL5A1 0.011 alpha 1 type V collagen preproprotein LMTK3 0.011 lemur tyrosine kinase 3 REXO1 0.011 transcription elongation factor B polypeptide 3 bin... COL3A1 0.010 collagen type III alpha 1 preproprotein NACA 0.010 nascent polypeptide-associated complex alpha subuni... CASKIN2 0.010 cask-interacting protein 2 isoform b CASKIN2 0.010 cask-interacting protein 2 isoform a KIAA1522 0.009 hypothetical protein LOC57648 SRRM1 0.009 serine/arginine repetitive matrix 1 COL11A2 0.009 collagen, type XI, alpha 2 isoform 2 preproprotein ... COL11A2 0.009 collagen, type XI, alpha 2 isoform 3 preproprotein ... COL11A2 0.009 collagen, type XI, alpha 2 isoform 1 preproprotein ... C7orf51 0.009 hypothetical protein FLJ37538 NEFH 0.008 neurofilament, heavy polypeptide 200kDa COL9A3 0.008 alpha 3 type IX collagen COL4A5 0.008 type IV collagen alpha 5 isoform 2 precursor COL4A5 0.008 type IV collagen alpha 5 isoform 1 precursorHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
