
| Name: TMEM131 | Sequence: fasta or formatted (1883aa) | NCBI GI: 150456424 | |
|
Description: RW1 protein
| Not currently referenced in the text | ||
|
Composition:
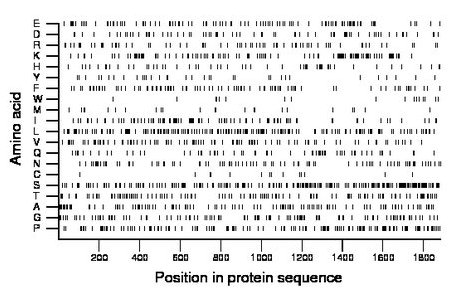
Amino acid Percentage Count Longest homopolymer A alanine 6.5 122 3 C cysteine 0.7 14 1 D aspartate 4.0 75 3 E glutamate 5.5 104 3 F phenylalanine 4.6 87 2 G glycine 6.3 118 3 H histidine 2.9 55 2 I isoleucine 5.4 101 3 K lysine 6.4 120 2 L leucine 8.8 166 2 M methionine 1.3 24 1 N asparagine 4.7 88 2 P proline 8.1 153 3 Q glutamine 3.4 64 2 R arginine 3.9 73 3 S serine 12.7 239 5 T threonine 6.5 123 3 V valine 5.5 104 2 W tryptophan 1.0 18 1 Y tyrosine 1.9 35 2 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 RW1 protein KIAA0922 0.061 hypothetical protein LOC23240 isoform 2 KIAA0922 0.061 hypothetical protein LOC23240 isoform 1 NOLC1 0.012 nucleolar and coiled-body phosphoprotein 1 NEFH 0.012 neurofilament, heavy polypeptide 200kDa PCLO 0.010 piccolo isoform 2 PCLO 0.010 piccolo isoform 1 TACC2 0.009 transforming, acidic coiled-coil containing protein ... MUC12 0.009 PREDICTED: mucin 12 BRD4 0.009 bromodomain-containing protein 4 isoform long POLR2A 0.009 DNA-directed RNA polymerase II A MLL2 0.008 myeloid/lymphoid or mixed-lineage leukemia 2 LOC100294236 0.008 PREDICTED: similar to diffuse panbronchiolitis crit... TACC2 0.008 transforming, acidic coiled-coil containing protein... TACC2 0.008 transforming, acidic coiled-coil containing protein ... TACC2 0.008 transforming, acidic coiled-coil containing protein ... DSPP 0.008 dentin sialophosphoprotein preproprotein ZC3H18 0.008 zinc finger CCCH-type containing 18 MICALL1 0.008 molecule interacting with Rab13 POM121C 0.008 POM121 membrane glycoprotein (rat)-like TRIM66 0.008 tripartite motif-containing 66 TNRC18 0.007 trinucleotide repeat containing 18 AFF1 0.007 myeloid/lymphoid or mixed-lineage leukemia trithorax ... AFF4 0.007 ALL1 fused gene from 5q31 SRRM1 0.007 serine/arginine repetitive matrix 1 DPCR1 0.007 diffuse panbronchiolitis critical region 1 protein ... PRG4 0.007 proteoglycan 4 isoform D PRG4 0.007 proteoglycan 4 isoform C RAPH1 0.007 Ras association and pleckstrin homology domains 1 is... POM121 0.007 nuclear pore membrane protein 121Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
