
| Name: MGC50722 | Sequence: fasta or formatted (953aa) | NCBI GI: 42733592 | |
|
Description: hypothetical protein LOC399693
| Not currently referenced in the text | ||
|
Composition:
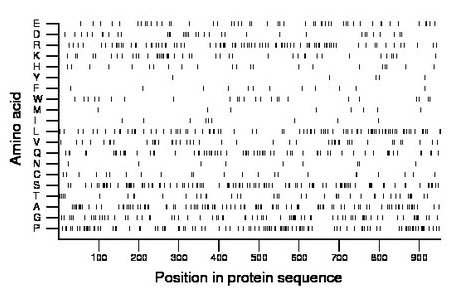
Amino acid Percentage Count Longest homopolymer A alanine 10.1 96 2 C cysteine 2.4 23 1 D aspartate 3.8 36 2 E glutamate 5.2 50 2 F phenylalanine 1.3 12 1 G glycine 8.0 76 2 H histidine 2.4 23 2 I isoleucine 0.9 9 1 K lysine 4.5 43 1 L leucine 8.9 85 3 M methionine 0.9 9 1 N asparagine 0.9 9 1 P proline 11.1 106 3 Q glutamine 7.1 68 3 R arginine 9.0 86 3 S serine 11.1 106 3 T threonine 4.3 41 2 V valine 4.8 46 2 W tryptophan 2.5 24 1 Y tyrosine 0.5 5 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 hypothetical protein LOC399693 CEP350 0.014 centrosome-associated protein 350 SRRM2 0.013 splicing coactivator subunit SRm300 SYNJ1 0.010 synaptojanin 1 isoform a SYNJ1 0.010 synaptojanin 1 isoform b OGFR 0.010 opioid growth factor receptor SCYL1 0.010 SCY1-like 1 isoform B MAP7D1 0.010 MAP7 domain containing 1 FLJ39639 0.009 PREDICTED: hypothetical protein FLJ39639 FLJ39639 0.009 PREDICTED: hypothetical protein FLJ39639 FLJ39639 0.009 PREDICTED: hypothetical protein LOC283876 SAMD1 0.009 sterile alpha motif domain containing 1 SYN1 0.009 synapsin I isoform Ia SRRM1 0.009 serine/arginine repetitive matrix 1 LOC100291634 0.009 PREDICTED: hypothetical protein XP_002346062 LOC100287232 0.009 PREDICTED: hypothetical protein XP_002342816 LOC100287232 0.009 PREDICTED: hypothetical protein SYNPO 0.008 synaptopodin isoform A USP36 0.008 ubiquitin specific peptidase 36 SYNJ1 0.008 synaptojanin 1 isoform d SYN1 0.008 synapsin I isoform Ib LOC729915 0.008 PREDICTED: similar to Putative POM121-like protein ... TCOF1 0.008 Treacher Collins-Franceschetti syndrome 1 isoform e... KNDC1 0.008 kinase non-catalytic C-lobe domain (KIND) containin... MDC1 0.007 mediator of DNA-damage checkpoint 1 FLJ31945 0.007 PREDICTED: hypothetical protein ANKRD33B 0.007 PREDICTED: ankyrin repeat domain 33B MAGI1 0.007 membrane associated guanylate kinase, WW and PDZ dom... NACA 0.007 nascent polypeptide-associated complex alpha subuni... LOC100292376 0.007 PREDICTED: hypothetical proteinHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
