
| Name: DBP | Sequence: fasta or formatted (325aa) | NCBI GI: 31542493 | |
|
Description: D site of albumin promoter (albumin D-box) binding protein
|
Referenced in:
| ||
|
Composition:
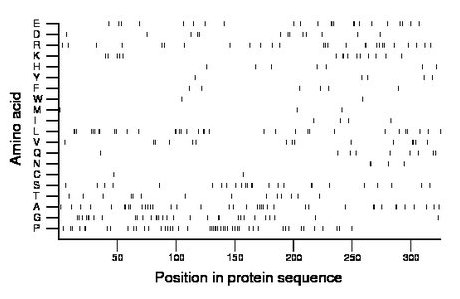
Amino acid Percentage Count Longest homopolymer A alanine 12.0 39 2 C cysteine 0.6 2 1 D aspartate 4.0 13 1 E glutamate 7.1 23 3 F phenylalanine 1.8 6 1 G glycine 9.5 31 3 H histidine 2.2 7 1 I isoleucine 1.2 4 1 K lysine 4.6 15 2 L leucine 10.5 34 3 M methionine 0.9 3 1 N asparagine 1.2 4 2 P proline 14.5 47 4 Q glutamine 2.8 9 1 R arginine 8.3 27 2 S serine 7.7 25 2 T threonine 4.3 14 2 V valine 4.6 15 2 W tryptophan 0.6 2 1 Y tyrosine 1.5 5 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 D site of albumin promoter (albumin D-box) binding p... HLF 0.350 hepatic leukemia factor TEF 0.308 thyrotrophic embryonic factor isoform 2 TEF 0.308 thyrotrophic embryonic factor isoform 1 LOC100291212 0.064 PREDICTED: hypothetical protein XP_002347943 NFIL3 0.053 nuclear factor, interleukin 3 regulated UNCX 0.050 UNC homeobox CEBPB 0.049 CCAAT/enhancer binding protein beta TPRX1 0.047 tetra-peptide repeat homeobox HCN2 0.046 hyperpolarization activated cyclic nucleotide-gated... EMID1 0.046 EMI domain containing 1 SFPQ 0.042 splicing factor proline/glutamine rich (polypyrimidin... CDKN1C 0.042 cyclin-dependent kinase inhibitor 1C isoform b [Hom... CDKN1C 0.042 cyclin-dependent kinase inhibitor 1C isoform b [Hom... CDKN1C 0.042 cyclin-dependent kinase inhibitor 1C isoform a CPSF6 0.041 cleavage and polyadenylation specific factor 6, 68 ... SGIP1 0.041 SH3-domain GRB2-like (endophilin) interacting prote... LOC440829 0.039 PREDICTED: transmembrane protein 46-like NACA 0.039 nascent polypeptide-associated complex alpha subuni... TAF4 0.038 TBP-associated factor 4 COL12A1 0.038 collagen, type XII, alpha 1 short isoform precursor ... COL12A1 0.038 collagen, type XII, alpha 1 long isoform precursor [... OGFR 0.038 opioid growth factor receptor PRG4 0.038 proteoglycan 4 isoform D PRG4 0.038 proteoglycan 4 isoform C PRG4 0.038 proteoglycan 4 isoform B PRG4 0.038 proteoglycan 4 isoform A SMARCC2 0.038 SWI/SNF-related matrix-associated actin-dependent r... SH3BP1 0.038 SH3-domain binding protein 1 KAT2A 0.036 general control of amino acid synthesis 5-like 2 [H...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
