
| Name: CEBPB | Sequence: fasta or formatted (345aa) | NCBI GI: 28872796 | |
|
Description: CCAAT/enhancer binding protein beta
|
Referenced in: Liver
| ||
|
Composition:
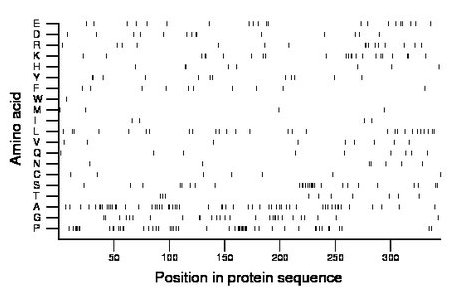
Amino acid Percentage Count Longest homopolymer A alanine 16.2 56 3 C cysteine 2.0 7 1 D aspartate 4.1 14 2 E glutamate 6.1 21 2 F phenylalanine 2.9 10 1 G glycine 8.1 28 3 H histidine 2.3 8 2 I isoleucine 1.2 4 1 K lysine 6.7 23 2 L leucine 8.1 28 2 M methionine 1.2 4 1 N asparagine 2.0 7 2 P proline 14.8 51 9 Q glutamine 2.6 9 1 R arginine 4.6 16 2 S serine 7.8 27 5 T threonine 2.9 10 1 V valine 2.6 9 1 W tryptophan 0.3 1 1 Y tyrosine 3.5 12 2 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 CCAAT/enhancer binding protein beta CEBPA 0.186 CCAAT/enhancer binding protein alpha CEBPE 0.163 CCAAT/enhancer binding protein epsilon CEBPD 0.158 CCAAT/enhancer binding protein delta CEBPG 0.078 CCAAT/enhancer binding protein gamma TAF4 0.046 TBP-associated factor 4 DBP 0.045 D site of albumin promoter (albumin D-box) binding p... FOXD1 0.042 forkhead box D1 ATF7 0.042 activating transcription factor 7 isoform 3 ATF7 0.042 activating transcription factor 7 isoform 2 ATF7 0.042 activating transcription factor 7 isoform 1 GRIN2D 0.041 N-methyl-D-aspartate receptor subunit 2D precursor ... SYNPO 0.041 synaptopodin isoform A SNX26 0.039 sorting nexin 26 LBXCOR1 0.036 LBXCOR1 homolog UNCX 0.036 UNC homeobox COL18A1 0.036 alpha 1 type XVIII collagen isoform 3 precursor [Ho... COL18A1 0.036 alpha 1 type XVIII collagen isoform 2 precursor [Ho... COL18A1 0.036 alpha 1 type XVIII collagen isoform 1 precursor [Ho... SALL3 0.036 sal-like 3 MLXIPL 0.035 Williams Beuren syndrome chromosome region 14 isofor... C12orf34 0.035 hypothetical protein LOC84915 MLXIPL 0.035 Williams Beuren syndrome chromosome region 14 isofor... ZIC5 0.035 zinc finger protein of the cerebellum 5 SHANK1 0.035 SH3 and multiple ankyrin repeat domains 1 MLXIPL 0.035 Williams Beuren syndrome chromosome region 14 isofor... MYPOP 0.035 Myb protein P42POP SP5 0.035 Sp5 transcription factor HCN4 0.033 hyperpolarization activated cyclic nucleotide-gated p... ZYX 0.033 zyxinHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
