
| Name: LOC100131988 | Sequence: fasta or formatted (234aa) | NCBI GI: 169204476 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
Other entries for this name:
alt mRNA {234aa} PREDICTED: hypothetical protein alt mRNA {234aa} PREDICTED: hypothetical protein | |||
|
Composition:
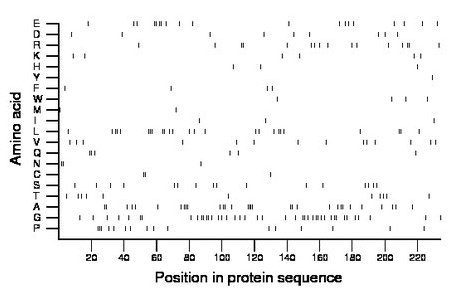
Amino acid Percentage Count Longest homopolymer A alanine 13.2 31 4 C cysteine 1.3 3 2 D aspartate 3.8 9 1 E glutamate 8.1 19 2 F phenylalanine 1.7 4 1 G glycine 17.9 42 2 H histidine 1.3 3 1 I isoleucine 1.3 3 1 K lysine 2.6 6 1 L leucine 10.7 25 3 M methionine 0.9 2 1 N asparagine 1.3 3 2 P proline 6.8 16 3 Q glutamine 2.6 6 2 R arginine 6.8 16 2 S serine 6.8 16 1 T threonine 4.7 11 1 V valine 6.0 14 1 W tryptophan 1.7 4 1 Y tyrosine 0.4 1 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC100131988 1.000 PREDICTED: hypothetical protein LOC100131988 1.000 PREDICTED: hypothetical protein ARID1B 0.038 AT rich interactive domain 1B (SWI1-like) isoform 2 ... ARID1B 0.038 AT rich interactive domain 1B (SWI1-like) isoform 1 ... ARID1B 0.038 AT rich interactive domain 1B (SWI1-like) isoform 3 ... IRS4 0.038 insulin receptor substrate 4 KRT2 0.038 keratin 2 AEBP2 0.038 AE binding protein 2 isoform b AEBP2 0.038 AE binding protein 2 isoform a KRT5 0.036 keratin 5 LOC100128960 0.033 PREDICTED: similar to paraneoplastic antigen MA2 [H... LOC100128960 0.033 PREDICTED: similar to Putative paraneoplastic antig... KRT9 0.031 keratin 9 AVEN 0.031 cell death regulator aven COL1A1 0.031 alpha 1 type I collagen preproprotein LOC100289969 0.031 PREDICTED: hypothetical protein XP_002347692 COL7A1 0.031 alpha 1 type VII collagen precursor TAF15 0.029 TBP-associated factor 15 isoform 2 TAF15 0.029 TBP-associated factor 15 isoform 1 LOC100290372 0.029 PREDICTED: hypothetical protein XP_002346788 LOC100288606 0.029 PREDICTED: hypothetical protein XP_002342652 EN2 0.029 engrailed homeobox 2 OSBP 0.029 oxysterol binding protein ONECUT3 0.029 one cut homeobox 3 BRWD3 0.029 bromodomain and WD repeat domain containing 3 [Homo... KHSRP 0.029 KH-type splicing regulatory protein FUS 0.029 fusion (involved in t(12;16) in malignant liposarcoma... DMKN 0.029 dermokine isoform 5 precursor DMKN 0.029 dermokine isoform 3 precursorHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
