
| Name: HRCT1 | Sequence: fasta or formatted (115aa) | NCBI GI: 89886261 | |
|
Description: histidine rich carboxyl terminus 1
| Not currently referenced in the text | ||
|
Composition:
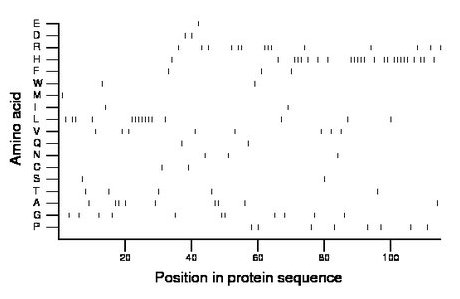
Amino acid Percentage Count Longest homopolymer A alanine 7.8 9 2 C cysteine 1.7 2 1 D aspartate 1.7 2 1 E glutamate 0.9 1 1 F phenylalanine 2.6 3 1 G glycine 9.6 11 2 H histidine 21.7 25 5 I isoleucine 1.7 2 1 K lysine 0.0 0 0 L leucine 13.0 15 7 M methionine 0.9 1 1 N asparagine 2.6 3 1 P proline 7.0 8 1 Q glutamine 1.7 2 1 R arginine 12.2 14 3 S serine 1.7 2 1 T threonine 4.3 5 1 V valine 7.0 8 1 W tryptophan 1.7 2 1 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 histidine rich carboxyl terminus 1 FOXB2 0.105 forkhead box B2 NLK 0.101 nemo like kinase OTX1 0.093 orthodenticle homeobox 1 HRG 0.089 histidine-rich glycoprotein precursor IQSEC1 0.084 IQ motif and Sec7 domain 1 isoform a NUFIP2 0.084 nuclear fragile X mental retardation protein interac... FOXG1 0.084 forkhead box G1 SIAH3 0.084 seven in absentia homolog 3 POU3F3 0.084 POU class 3 homeobox 3 DYRK1A 0.080 dual-specificity tyrosine-(Y)-phosphorylation regula... DYRK1A 0.080 dual-specificity tyrosine-(Y)-phosphorylation regula... SLC39A6 0.080 solute carrier family 39 (zinc transporter), member... SLC39A6 0.080 solute carrier family 39 (zinc transporter), member... MAFA 0.080 v-maf musculoaponeurotic fibrosarcoma oncogene homol... CREB5 0.076 cAMP responsive element binding protein 5 isoform de... CREB5 0.076 cAMP responsive element binding protein 5 isoform be... CREB5 0.076 cAMP responsive element binding protein 5 isoform ga... CREB5 0.076 cAMP responsive element binding protein 5 isoform al... ARID1B 0.076 AT rich interactive domain 1B (SWI1-like) isoform 2 ... ARID1B 0.076 AT rich interactive domain 1B (SWI1-like) isoform 1 ... ARID1B 0.076 AT rich interactive domain 1B (SWI1-like) isoform 3 ... VGLL3 0.076 colon carcinoma related protein C10orf140 0.072 hypothetical protein LOC387640 NKD2 0.072 naked cuticle homolog 2 CBX4 0.068 chromobox homolog 4 ONECUT2 0.068 one cut domain, family member 2 GSX2 0.068 GS homeobox 2 DLGAP3 0.063 discs, large (Drosophila) homolog-associated protei... CHD8 0.063 chromodomain helicase DNA binding protein 8Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
