
| Name: ZBED3 | Sequence: fasta or formatted (234aa) | NCBI GI: 14150185 | |
|
Description: zinc finger, BED-type containing 3
|
Referenced in: Small RNAs in RNA Processing
| ||
|
Composition:
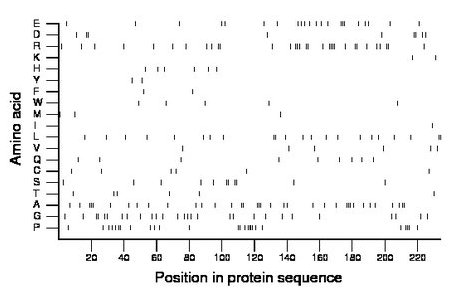
Amino acid Percentage Count Longest homopolymer A alanine 15.4 36 3 C cysteine 2.6 6 1 D aspartate 3.8 9 2 E glutamate 9.4 22 3 F phenylalanine 0.9 2 1 G glycine 12.0 28 2 H histidine 2.6 6 1 I isoleucine 0.4 1 1 K lysine 0.9 2 1 L leucine 9.4 22 2 M methionine 1.3 3 1 N asparagine 0.0 0 0 P proline 11.1 26 4 Q glutamine 3.8 9 1 R arginine 13.7 32 4 S serine 4.7 11 2 T threonine 2.6 6 1 V valine 2.6 6 1 W tryptophan 2.1 5 1 Y tyrosine 0.9 2 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 zinc finger, BED-type containing 3 ZBED2 0.316 zinc finger, BED domain containing 2 UNCX 0.045 UNC homeobox TBC1D10B 0.043 TBC1 domain family, member 10B BAT2 0.041 HLA-B associated transcript-2 LOC100130174 0.041 PREDICTED: hypothetical protein LOC100130174 0.041 PREDICTED: hypothetical protein LOC100130174 0.041 PREDICTED: hypothetical protein TCHH 0.041 trichohyalin SCRIB 0.039 scribble isoform b SCRIB 0.039 scribble isoform a UBXN1 0.039 UBX domain protein 1 CCDC102A 0.039 coiled-coil domain containing 102A ACIN1 0.039 apoptotic chromatin condensation inducer 1 CEBPD 0.039 CCAAT/enhancer binding protein delta MAP7D1 0.039 MAP7 domain containing 1 CALD1 0.037 caldesmon 1 isoform 1 LOC441086 0.034 PREDICTED: hypothetical protein FBXO41 0.034 F-box protein 41 LOC100287514 0.034 PREDICTED: hypothetical protein XP_002343754 KIAA1211 0.032 hypothetical protein LOC57482 LOC441086 0.032 PREDICTED: hypothetical protein EMILIN1 0.032 elastin microfibril interfacer 1 CDC2L1 0.032 cell division cycle 2-like 1 (PITSLRE proteins) isof... ZBED4 0.032 zinc finger, BED-type containing 4 ANKRD24 0.032 ankyrin repeat domain 24 SYNPO 0.030 synaptopodin isoform A CABP1 0.030 calcium binding protein 1 isoform 3 LOC100289979 0.030 PREDICTED: hypothetical protein XP_002346878 LOC100289273 0.030 PREDICTED: hypothetical protein XP_002342720Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
