
| Name: WBSCR16 | Sequence: fasta or formatted (464aa) | NCBI GI: 13540582 | |
|
Description: Williams-Beuren syndrome chromosome region 16
|
Referenced in:
| ||
|
Composition:
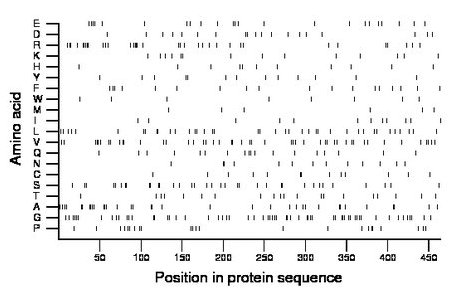
Amino acid Percentage Count Longest homopolymer A alanine 7.5 35 2 C cysteine 2.8 13 2 D aspartate 4.1 19 1 E glutamate 5.0 23 1 F phenylalanine 3.9 18 1 G glycine 12.9 60 2 H histidine 2.4 11 1 I isoleucine 2.6 12 1 K lysine 3.0 14 1 L leucine 8.4 39 2 M methionine 1.5 7 1 N asparagine 2.8 13 2 P proline 5.0 23 2 Q glutamine 3.9 18 1 R arginine 7.5 35 3 S serine 7.5 35 2 T threonine 4.3 20 1 V valine 10.3 48 2 W tryptophan 1.7 8 1 Y tyrosine 2.8 13 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 Williams-Beuren syndrome chromosome region 16 LOC653375 0.577 PREDICTED: RCC1-like G exchanging factor-like [Homo... LOC653375 0.577 PREDICTED: RCC1-like G exchanging factor-like [Homo... HERC3 0.104 hect domain and RLD 3 HERC4 0.102 hect domain and RLD 4 isoform b HERC4 0.102 hect domain and RLD 4 isoform a HERC6 0.086 hect domain and RLD 6 HERC1 0.083 guanine nucleotide exchange factor p532 RCBTB2 0.081 regulator of chromosome condensation and BTB domain c... RCBTB1 0.073 regulator of chromosome condensation (RCC1) and BTB ... HERC2 0.070 hect domain and RLD 2 HERC5 0.070 hect domain and RLD 5 RCC1 0.067 regulator of chromosome condensation 1 isoform b [H... RCC1 0.067 regulator of chromosome condensation 1 isoform a [H... RCC1 0.067 regulator of chromosome condensation 1 isoform c [Hom... SNHG3-RCC1 0.067 regulator of chromosome condensation 1 SNHG3-RCC1 0.067 regulator of chromosome condensation 1 SNHG3-RCC1 0.067 regulator of chromosome condensation 1 RPGR 0.064 retinitis pigmentosa GTPase regulator isoform A [Homo... RPGR 0.064 retinitis pigmentosa GTPase regulator isoform C [Hom... RCC2 0.061 regulator of chromosome condensation 2 RCC2 0.061 regulator of chromosome condensation 2 SERGEF 0.059 deafness locus associated putative guanine nucleotide... NEK9 0.050 NIMA-related kinase 9 NEK8 0.042 NIMA-related kinase 8 RCCD1 0.040 RCC1 domain containing 1 RCCD1 0.040 RCC1 domain containing 1 IBTK 0.037 inhibitor of Bruton's tyrosine kinase LOC100290629 0.031 PREDICTED: hypothetical protein XP_002347563 LOC390561 0.030 PREDICTED: similar to hect domain and RLD 2Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
