
| Name: LOC100292187 | Sequence: fasta or formatted (202aa) | NCBI GI: 239757829 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
|
Composition:
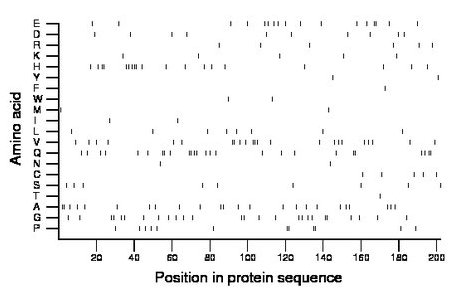
Amino acid Percentage Count Longest homopolymer A alanine 11.9 24 2 C cysteine 2.5 5 1 D aspartate 5.0 10 1 E glutamate 8.4 17 2 F phenylalanine 0.5 1 1 G glycine 12.9 26 2 H histidine 9.4 19 3 I isoleucine 1.0 2 1 K lysine 2.5 5 1 L leucine 4.0 8 1 M methionine 1.0 2 1 N asparagine 1.0 2 1 P proline 5.9 12 2 Q glutamine 12.9 26 2 R arginine 3.0 6 1 S serine 4.5 9 1 T threonine 0.5 1 1 V valine 11.4 23 3 W tryptophan 1.0 2 1 Y tyrosine 1.0 2 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC100289845 0.965 PREDICTED: hypothetical protein XP_002348205 LOC100289046 0.965 PREDICTED: hypothetical protein XP_002343864 LOC100292515 0.060 PREDICTED: hypothetical protein LOC100289910 0.060 PREDICTED: hypothetical protein XP_002347211 LOC100287264 0.060 PREDICTED: hypothetical protein XP_002343075 LOC100291688 0.042 PREDICTED: hypothetical protein XP_002345241 LOC100290054 0.042 PREDICTED: hypothetical protein XP_002348009 LOC100287338 0.042 PREDICTED: hypothetical protein XP_002343730 LOC100292013 0.037 PREDICTED: similar to LMARRLC1A LOC100287831 0.037 PREDICTED: similar to LMARRLC1A LOC100287831 0.037 PREDICTED: hypothetical protein XP_002343735 FUSSEL18 0.027 PREDICTED: functional smad suppressing element 18 [... FUSSEL18 0.027 PREDICTED: functional smad suppressing element 18 [... FUSSEL18 0.027 PREDICTED: functional smad suppressing element 18 [... HOXA1 0.027 homeobox A1 isoform a HOXA1 0.027 homeobox A1 isoform b LOC100292716 0.025 PREDICTED: similar to pseudoautosomal GTP-binding p... FOXG1 0.022 forkhead box G1 CACNA1A 0.022 calcium channel, alpha 1A subunit isoform 4 CACNA1A 0.022 calcium channel, alpha 1A subunit isoform 3 ARID1B 0.022 AT rich interactive domain 1B (SWI1-like) isoform 2 ... ARID1B 0.022 AT rich interactive domain 1B (SWI1-like) isoform 1 ... ARID1B 0.022 AT rich interactive domain 1B (SWI1-like) isoform 3 ... MAFB 0.020 transcription factor MAFB DYRK1A 0.020 dual-specificity tyrosine-(Y)-phosphorylation regula... DYRK1A 0.020 dual-specificity tyrosine-(Y)-phosphorylation regula... NLK 0.020 nemo like kinase DLX6 0.017 distal-less homeobox 6 GSX2 0.017 GS homeobox 2Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
