
| Name: LOC100292013 | Sequence: fasta or formatted (279aa) | NCBI GI: 239757352 | |
|
Description: PREDICTED: similar to LMARRLC1A
| Not currently referenced in the text | ||
|
Composition:
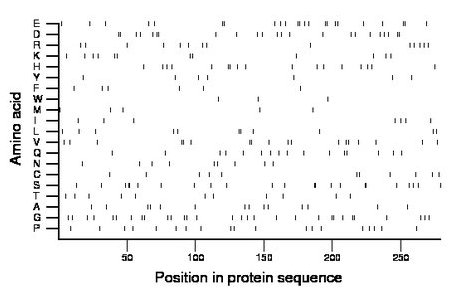
Amino acid Percentage Count Longest homopolymer A alanine 5.0 14 1 C cysteine 3.9 11 1 D aspartate 8.2 23 2 E glutamate 8.2 23 2 F phenylalanine 2.2 6 1 G glycine 10.8 30 2 H histidine 6.1 17 2 I isoleucine 2.5 7 1 K lysine 4.3 12 1 L leucine 3.9 11 2 M methionine 1.8 5 1 N asparagine 3.2 9 1 P proline 6.8 19 1 Q glutamine 5.7 16 1 R arginine 5.0 14 1 S serine 8.2 23 2 T threonine 3.9 11 1 V valine 6.5 18 2 W tryptophan 1.1 3 1 Y tyrosine 2.5 7 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: similar to LMARRLC1A LOC100287831 0.997 PREDICTED: similar to LMARRLC1A LOC100287831 0.991 PREDICTED: hypothetical protein XP_002343735 LOC100287311 0.059 PREDICTED: hypothetical protein XP_002343729 LOC100291688 0.059 PREDICTED: hypothetical protein XP_002345241 LOC100290054 0.059 PREDICTED: hypothetical protein XP_002348009 LOC100287338 0.026 PREDICTED: hypothetical protein XP_002343730 LOC100292187 0.026 PREDICTED: hypothetical protein LOC100289845 0.026 PREDICTED: hypothetical protein XP_002348205 LOC100289046 0.026 PREDICTED: hypothetical protein XP_002343864 LOC100132713 0.026 PREDICTED: similar to hCG1654128 LOC653203 0.024 PREDICTED: hypothetical protein LOC100132713 0.021 PREDICTED: similar to hCG1654128 LOC100132713 0.021 PREDICTED: similar to hCG1654128 LOC727962 0.019 PREDICTED: similar to collagen, type I, alpha 2 [Ho... HRNR 0.014 hornerin LRIG1 0.014 leucine-rich repeats and immunoglobulin-like domains... HEG1 0.014 HEG homolog 1 ACBD3 0.014 acyl-Coenzyme A binding domain containing 3 NISCH 0.012 nischarin SI 0.009 sucrase-isomaltase HRC 0.009 histidine rich calcium binding protein precursor [Hom... BMP2K 0.009 BMP-2 inducible kinase isoform a TNFRSF8 0.009 tumor necrosis factor receptor superfamily, member 8... SCN4A 0.009 voltage-gated sodium channel type 4 alpha SEC16A 0.009 SEC16 homolog A SUPT7L 0.009 SPTF-associated factor 65 gamma CD177 0.009 CD177 molecule ILDR2 0.009 immunoglobulin-like domain containing receptor 2 [Ho... OBSCN 0.007 obscurin, cytoskeletal calmodulin and titin-interac...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
