
| Name: LOC100287616 | Sequence: fasta or formatted (122aa) | NCBI GI: 239745230 | |
|
Description: PREDICTED: hypothetical protein XP_002343393
| Not currently referenced in the text | ||
|
Composition:
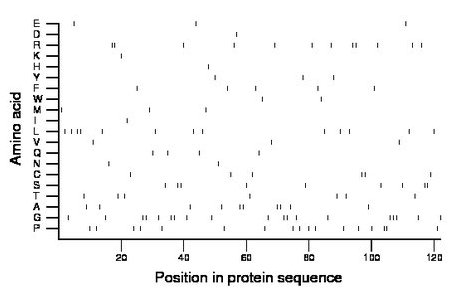
Amino acid Percentage Count Longest homopolymer A alanine 8.2 10 2 C cysteine 4.9 6 2 D aspartate 0.8 1 1 E glutamate 2.5 3 1 F phenylalanine 4.1 5 1 G glycine 15.6 19 3 H histidine 0.8 1 1 I isoleucine 0.8 1 1 K lysine 0.8 1 1 L leucine 10.7 13 2 M methionine 2.5 3 1 N asparagine 1.6 2 1 P proline 13.9 17 2 Q glutamine 3.3 4 1 R arginine 9.8 12 2 S serine 7.4 9 2 T threonine 5.7 7 1 V valine 2.5 3 1 W tryptophan 1.6 2 1 Y tyrosine 2.5 3 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002343393 LOC100292820 0.469 PREDICTED: hypothetical protein KDM6B 0.041 lysine (K)-specific demethylase 6B LOC728431 0.029 PREDICTED: hypothetical protein ZNF469 0.021 zinc finger protein 469 LOC100292534 0.021 PREDICTED: hypothetical protein LOC100290655 0.021 PREDICTED: hypothetical protein XP_002347968 LOC100287933 0.021 PREDICTED: hypothetical protein XP_002343257 LOC100134152 0.017 PREDICTED: similar to synaptotagmin XV-b CACNA2D2 0.017 calcium channel, voltage-dependent, alpha 2/delta su... CACNA2D2 0.017 calcium channel, voltage-dependent, alpha 2/delta su... C1QTNF5 0.017 C1q and tumor necrosis factor related protein 5 prec... NISCH 0.017 nischarin NOTCH1 0.012 notch1 preproprotein LOC100288150 0.012 PREDICTED: hypothetical protein XP_002343661 LOC100289575 0.012 PREDICTED: hypothetical protein XP_002342245 POU5F2 0.012 POU domain class 5, transcription factor 2 LRP8 0.012 low density lipoprotein receptor-related protein 8 i... TFAP2E 0.012 transcription factor AP-2 epsilon (activating enhan... MSX1 0.012 msh homeobox 1 LOC283999 0.008 hypothetical protein LOC283999 AGAP2 0.008 centaurin, gamma 1 isoform PIKE-L SQSTM1 0.008 sequestosome 1 isoform 2 SQSTM1 0.008 sequestosome 1 isoform 2 SQSTM1 0.008 sequestosome 1 isoform 1 BBC3 0.008 BCL2 binding component 3 isoform 1 DIDO1 0.008 death inducer-obliterator 1 isoform c C17orf102 0.008 hypothetical protein LOC400591 LOC100292408 0.008 PREDICTED: hypothetical protein LOC100291136 0.008 PREDICTED: hypothetical protein XP_002347630Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
