
| Name: ZSWIM5 | Sequence: fasta or formatted (1185aa) | NCBI GI: 149944451 | |
|
Description: zinc finger, SWIM domain containing 5
|
Referenced in:
| ||
|
Composition:
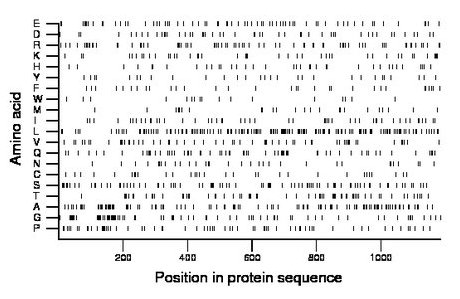
Amino acid Percentage Count Longest homopolymer A alanine 9.5 113 3 C cysteine 2.6 31 1 D aspartate 4.2 50 2 E glutamate 6.1 72 2 F phenylalanine 2.6 31 1 G glycine 6.8 81 3 H histidine 3.2 38 2 I isoleucine 4.3 51 2 K lysine 3.6 43 3 L leucine 12.7 150 3 M methionine 2.1 25 1 N asparagine 2.7 32 1 P proline 5.6 66 3 Q glutamine 5.5 65 3 R arginine 6.2 74 3 S serine 7.6 90 3 T threonine 5.7 67 3 V valine 5.1 60 2 W tryptophan 1.4 17 1 Y tyrosine 2.4 29 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 zinc finger, SWIM domain containing 5 ZSWIM6 0.720 zinc finger, SWIM-type containing 6 ZSWIM4 0.341 zinc finger, SWIM-type containing 4 KIAA0913 0.066 hypothetical protein LOC23053 GSK3A 0.010 glycogen synthase kinase 3 alpha COL10A1 0.008 type X collagen alpha 1 precursor COL1A1 0.008 alpha 1 type I collagen preproprotein VASP 0.008 vasodilator-stimulated phosphoprotein KHSRP 0.008 KH-type splicing regulatory protein MAP3K12 0.008 mitogen-activated protein kinase kinase kinase 12 [H... SPRN 0.008 shadow of prion protein POU4F1 0.008 POU domain, class 4, transcription factor 1 NOVA2 0.008 neuro-oncological ventral antigen 2 PHOX2B 0.008 paired-like homeobox 2b C6orf174 0.008 hypothetical protein LOC387104 ELN 0.007 elastin isoform e precursor ELN 0.007 elastin isoform d precursor ELN 0.007 elastin isoform c precursor ELN 0.007 elastin isoform b precursor ELN 0.007 elastin isoform a precursor HOXD11 0.007 homeobox D11 TAF4 0.007 TBP-associated factor 4 KAT2B 0.007 K(lysine) acetyltransferase 2B KLF16 0.007 BTE-binding protein 4 HOXA10 0.007 homeobox A10 isoform a RANBP9 0.007 RAN binding protein 9 BHLHE22 0.007 basic helix-loop-helix domain containing, class B, 5... LTK 0.007 leukocyte receptor tyrosine kinase isoform 1 precurs... FOXD1 0.007 forkhead box D1 UNCX 0.007 UNC homeoboxHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
