
| Name: FLG | Sequence: fasta or formatted (4061aa) | NCBI GI: 60097902 | |
|
Description: filaggrin
|
Referenced in: Calmodulin and Calcium
| ||
|
Composition:
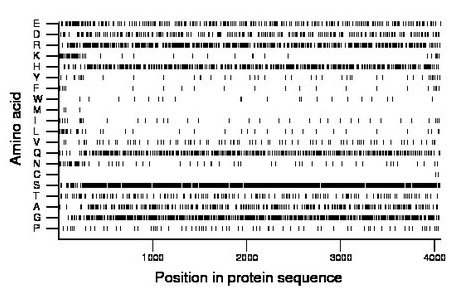
Amino acid Percentage Count Longest homopolymer A alanine 6.1 246 2 C cysteine 0.0 2 1 D aspartate 5.5 222 2 E glutamate 6.3 256 3 F phenylalanine 0.6 24 1 G glycine 12.8 518 2 H histidine 10.2 413 3 I isoleucine 0.9 38 2 K lysine 1.4 57 2 L leucine 1.1 43 2 M methionine 0.1 5 1 N asparagine 1.5 62 2 P proline 1.7 69 1 Q glutamine 9.0 367 2 R arginine 10.8 440 2 S serine 24.1 977 4 T threonine 4.1 166 2 V valine 2.0 83 2 W tryptophan 0.5 21 1 Y tyrosine 1.3 52 4 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 filaggrin HRNR 0.104 hornerin FLG2 0.089 filaggrin family member 2 DSPP 0.029 dentin sialophosphoprotein preproprotein MUC12 0.024 PREDICTED: mucin 12 SRRM2 0.019 splicing coactivator subunit SRm300 RPTN 0.018 repetin RBMXL3 0.015 RNA binding motif protein, X-linked-like 3 TCHH 0.012 trichohyalin NKTR 0.012 natural killer-tumor recognition sequence MUC12 0.011 PREDICTED: mucin 12, cell surface associated ZC3H13 0.010 zinc finger CCCH-type containing 13 CRNN 0.010 cornulin LOC100292370 0.009 PREDICTED: hypothetical protein GPATCH8 0.009 G patch domain containing 8 LOC100170229 0.009 hypothetical protein LOC100170229 C2orf16 0.009 hypothetical protein LOC84226 HRC 0.009 histidine rich calcium binding protein precursor [Hom... DMP1 0.008 dentin matrix acidic phosphoprotein 1 isoform 1 precu... MUC21 0.008 mucin 21 TCHHL1 0.008 trichohyalin-like 1 RP1L1 0.008 retinitis pigmentosa 1-like 1 KIAA1853 0.008 KIAA1853 protein MUC12 0.008 PREDICTED: mucin 12 RPGR 0.008 retinitis pigmentosa GTPase regulator isoform C [Hom... FLJ37078 0.008 hypothetical protein LOC222183 MED1 0.007 mediator complex subunit 1 AFF4 0.007 ALL1 fused gene from 5q31 TCOF1 0.007 Treacher Collins-Franceschetti syndrome 1 isoform e... SFRS4 0.007 splicing factor, arginine/serine-rich 4Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
