
| Name: PTMS | Sequence: fasta or formatted (102aa) | NCBI GI: 46276863 | |
|
Description: parathymosin
|
Referenced in:
| ||
|
Composition:
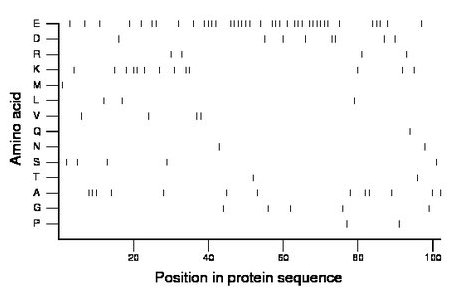
Amino acid Percentage Count Longest homopolymer A alanine 12.7 13 3 C cysteine 0.0 0 0 D aspartate 7.8 8 2 E glutamate 38.2 39 6 F phenylalanine 0.0 0 0 G glycine 4.9 5 1 H histidine 0.0 0 0 I isoleucine 0.0 0 0 K lysine 12.7 13 2 L leucine 2.9 3 1 M methionine 1.0 1 1 N asparagine 2.0 2 1 P proline 2.0 2 1 Q glutamine 1.0 1 1 R arginine 3.9 4 1 S serine 4.9 5 1 T threonine 2.0 2 1 V valine 3.9 4 2 W tryptophan 0.0 0 0 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 parathymosin PTMA 0.329 prothymosin, alpha isoform 2 PTMA 0.329 prothymosin, alpha isoform 1 LOC441454 0.329 PREDICTED: hypothetical protein LOC728026 0.318 PREDICTED: hypothetical protein LOC728026 0.318 PREDICTED: hypothetical protein LOC728026 0.318 PREDICTED: hypothetical protein MYT1 0.231 myelin transcription factor 1 RPGR 0.225 retinitis pigmentosa GTPase regulator isoform C [Hom... LOC100294088 0.214 PREDICTED: hypothetical protein, partial YTHDC1 0.202 splicing factor YT521-B isoform 2 YTHDC1 0.202 splicing factor YT521-B isoform 1 PELP1 0.202 proline, glutamic acid and leucine rich protein 1 [... LOC100289089 0.197 PREDICTED: hypothetical protein XP_002343359 LOC728792 0.191 PREDICTED: hypothetical protein WDR87 0.185 NYD-SP11 protein NEFL 0.185 neurofilament, light polypeptide 68kDa NEFM 0.185 neurofilament, medium polypeptide 150kDa isoform 1 ... NEFM 0.185 neurofilament, medium polypeptide 150kDa isoform 2 ... SUPT5H 0.185 suppressor of Ty 5 homolog isoform b SUPT5H 0.185 suppressor of Ty 5 homolog isoform a SUPT5H 0.185 suppressor of Ty 5 homolog isoform a SUPT5H 0.185 suppressor of Ty 5 homolog isoform a EIF5B 0.185 eukaryotic translation initiation factor 5B FAM9A 0.185 family with sequence similarity 9, member A LOXHD1 0.179 lipoxygenase homology domains 1 isoform 1 HMGB1 0.179 high-mobility group box 1 RGAG4 0.179 retrotransposon gag domain containing 4 CENPB 0.173 centromere protein B NEFH 0.173 neurofilament, heavy polypeptide 200kDaHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
