
| Name: PTMA | Sequence: fasta or formatted (111aa) | NCBI GI: 151101407 | |
|
Description: prothymosin, alpha isoform 1
|
Referenced in:
| ||
Other entries for this name:
alt prot [110aa] prothymosin, alpha isoform 2 | |||
|
Composition:
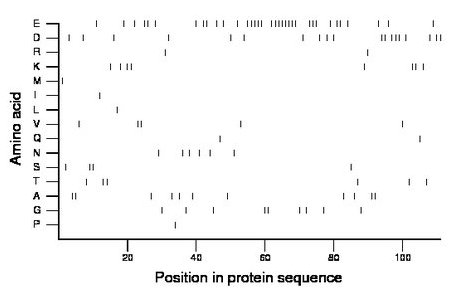
Amino acid Percentage Count Longest homopolymer A alanine 9.9 11 2 C cysteine 0.0 0 0 D aspartate 17.1 19 3 E glutamate 31.5 35 8 F phenylalanine 0.0 0 0 G glycine 8.1 9 2 H histidine 0.0 0 0 I isoleucine 0.9 1 1 K lysine 7.2 8 2 L leucine 0.9 1 1 M methionine 0.9 1 1 N asparagine 5.4 6 1 P proline 0.9 1 1 Q glutamine 1.8 2 1 R arginine 1.8 2 1 S serine 3.6 4 2 T threonine 5.4 6 2 V valine 4.5 5 2 W tryptophan 0.0 0 0 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 prothymosin, alpha isoform 1 PTMA 0.970 prothymosin, alpha isoform 2 LOC728026 0.859 PREDICTED: hypothetical protein LOC728026 0.859 PREDICTED: hypothetical protein LOC728026 0.859 PREDICTED: hypothetical protein LOC441454 0.854 PREDICTED: hypothetical protein PTMS 0.288 parathymosin MYT1 0.197 myelin transcription factor 1 UBTF 0.187 upstream binding transcription factor, RNA polymera... UBTF 0.187 upstream binding transcription factor, RNA polymera... UBTF 0.187 upstream binding transcription factor, RNA polymerase... ANP32B 0.182 acidic (leucine-rich) nuclear phosphoprotein 32 famil... YTHDC1 0.182 splicing factor YT521-B isoform 2 YTHDC1 0.182 splicing factor YT521-B isoform 1 LOC728792 0.177 PREDICTED: hypothetical protein TRIM44 0.177 DIPB protein RPGR 0.177 retinitis pigmentosa GTPase regulator isoform C [Hom... LOC647020 0.177 PREDICTED: similar to acidic (leucine-rich) nuclear... LOC647020 0.177 PREDICTED: similar to acidic (leucine-rich) nuclear... MYT1L 0.177 myelin transcription factor 1-like PELP1 0.172 proline, glutamic acid and leucine rich protein 1 [... NEFM 0.172 neurofilament, medium polypeptide 150kDa isoform 1 ... NEFM 0.172 neurofilament, medium polypeptide 150kDa isoform 2 ... NCL 0.167 nucleolin CENPB 0.162 centromere protein B HUWE1 0.162 HECT, UBA and WWE domain containing 1 NEFL 0.157 neurofilament, light polypeptide 68kDa CDC2L2 0.157 cell division cycle 2-like 2 isoform 4 CDC2L2 0.157 cell division cycle 2-like 2 isoform 1 CDC2L1 0.157 cell division cycle 2-like 1 (PITSLRE proteins) isof...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
