
| Name: TIGD1 | Sequence: fasta or formatted (591aa) | NCBI GI: 22209001 | |
|
Description: hypothetical protein LOC200765
|
Referenced in: DNA Transposon and Retrovirus-related Sequences
| ||
|
Composition:
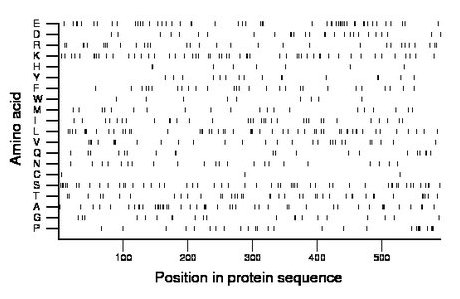
Amino acid Percentage Count Longest homopolymer A alanine 8.1 48 3 C cysteine 0.5 3 1 D aspartate 4.6 27 2 E glutamate 9.0 53 2 F phenylalanine 4.6 27 1 G glycine 3.6 21 2 H histidine 1.4 8 1 I isoleucine 5.4 32 2 K lysine 9.0 53 2 L leucine 9.0 53 3 M methionine 3.6 21 1 N asparagine 3.7 22 1 P proline 4.4 26 3 Q glutamine 3.4 20 2 R arginine 5.2 31 2 S serine 8.8 52 2 T threonine 6.8 40 2 V valine 4.9 29 2 W tryptophan 1.5 9 1 Y tyrosine 2.7 16 2 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 hypothetical protein LOC200765 TIGD2 0.171 tigger transposable element derived 2 JRKL 0.169 jerky homolog-like LOC100132176 0.159 PREDICTED: similar to HDCMB45P TIGD7 0.135 tigger transposable element derived 7 JRK 0.127 jerky isoform a JRK 0.127 jerky isoform b TIGD6 0.119 hypothetical protein LOC81789 TIGD4 0.098 tigger transposable element derived 4 TIGD5 0.086 tigger transposable element derived 5 MGC16385 0.070 hypothetical protein LOC92806 CENPB 0.061 centromere protein B DCHS2 0.050 dachsous 2 isoform 1 TIGD3 0.041 tigger transposable element derived 3 LOC100293487 0.032 PREDICTED: similar to cag LOC100291289 0.032 PREDICTED: hypothetical protein XP_002347296 LOC100287896 0.032 PREDICTED: hypothetical protein XP_002343159 WDR67 0.008 WD repeat domain 67 isoform 1 LOC100293326 0.007 PREDICTED: similar to hCG1802057 ANKRD18B 0.007 PREDICTED: ankyrin repeat domain 18B LOC100291015 0.007 PREDICTED: hypothetical protein XP_002347719 ANKRD18B 0.007 PREDICTED: ankyrin repeat domain 18B LOC100288326 0.007 PREDICTED: hypothetical protein XP_002344143 ANKRD18B 0.007 PREDICTED: ankyrin repeat domain 18B ANKRD18A 0.007 PREDICTED: ankyrin repeat domain 18A ANKRD18A 0.007 PREDICTED: ankyrin repeat domain 18A ANKRD18A 0.007 PREDICTED: ankyrin repeat domain 18A MYH7B 0.007 myosin, heavy polypeptide 7B, cardiac muscle, beta ... SYNE1 0.006 spectrin repeat containing, nuclear envelope 1 isofo... SYNE1 0.006 spectrin repeat containing, nuclear envelope 1 isof...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
