
| Name: PRR7 | Sequence: fasta or formatted (274aa) | NCBI GI: 21361937 | |
|
Description: proline rich 7 (synaptic)
|
Referenced in:
| ||
|
Composition:
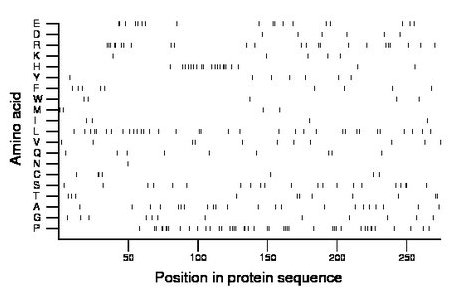
Amino acid Percentage Count Longest homopolymer A alanine 8.0 22 1 C cysteine 2.6 7 2 D aspartate 2.2 6 1 E glutamate 6.6 18 2 F phenylalanine 3.3 9 1 G glycine 4.4 12 1 H histidine 7.3 20 3 I isoleucine 1.5 4 1 K lysine 1.8 5 1 L leucine 12.4 34 2 M methionine 1.5 4 1 N asparagine 0.4 1 1 P proline 16.1 44 4 Q glutamine 3.3 9 1 R arginine 8.8 24 3 S serine 7.7 21 2 T threonine 4.0 11 2 V valine 4.0 11 1 W tryptophan 1.8 5 1 Y tyrosine 2.6 7 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 proline rich 7 (synaptic) LOC100288473 0.060 PREDICTED: hypothetical protein XP_002343336 LOC100293557 0.056 PREDICTED: hypothetical protein PHLDA1 0.055 pleckstrin homology-like domain, family A, member 1 ... MAFB 0.049 transcription factor MAFB LOC100131525 0.049 PREDICTED: hypothetical protein FOXB2 0.049 forkhead box B2 RBM33 0.046 RNA binding motif protein 33 CREB5 0.044 cAMP responsive element binding protein 5 isoform de... CREB5 0.044 cAMP responsive element binding protein 5 isoform be... CREB5 0.044 cAMP responsive element binding protein 5 isoform ga... CREB5 0.044 cAMP responsive element binding protein 5 isoform al... RNF111 0.042 ring finger protein 111 LOC100291393 0.041 PREDICTED: hypothetical protein XP_002347924 SF3B4 0.041 splicing factor 3b, subunit 4 FUSSEL18 0.039 PREDICTED: functional smad suppressing element 18 [... FUSSEL18 0.039 PREDICTED: functional smad suppressing element 18 [... FUSSEL18 0.039 PREDICTED: functional smad suppressing element 18 [... LOC100288786 0.039 PREDICTED: hypothetical protein XP_002343819 MLL5 0.037 myeloid/lymphoid or mixed-lineage leukemia 5 MLL5 0.037 myeloid/lymphoid or mixed-lineage leukemia 5 PDE4D 0.035 phosphodiesterase 4D isoform 1 BRD4 0.035 bromodomain-containing protein 4 isoform long SHANK1 0.035 SH3 and multiple ankyrin repeat domains 1 WIPF2 0.035 WIRE protein FOXL2 0.034 forkhead box L2 FLJ22184 0.034 PREDICTED: hypothetical protein FLJ22184 C10orf140 0.034 hypothetical protein LOC387640 FOXJ3 0.034 forkhead box J3 SF1 0.034 splicing factor 1 isoform 2Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
