
| Name: BCL7B | Sequence: fasta or formatted (202aa) | NCBI GI: 20336473 | |
|
Description: B-cell CLL/lymphoma 7B
|
Referenced in:
| ||
|
Composition:
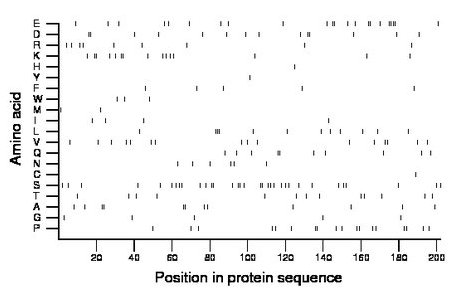
Amino acid Percentage Count Longest homopolymer A alanine 5.9 12 2 C cysteine 0.5 1 1 D aspartate 6.9 14 2 E glutamate 10.9 22 4 F phenylalanine 2.5 5 1 G glycine 2.5 5 1 H histidine 0.5 1 1 I isoleucine 2.0 4 1 K lysine 7.4 15 2 L leucine 5.9 12 3 M methionine 1.0 2 1 N asparagine 3.0 6 1 P proline 8.9 18 2 Q glutamine 5.0 10 2 R arginine 4.5 9 1 S serine 16.3 33 2 T threonine 6.4 13 1 V valine 7.9 16 2 W tryptophan 1.5 3 1 Y tyrosine 0.5 1 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 B-cell CLL/lymphoma 7B BCL7C 0.393 B-cell CLL/lymphoma 7C BCL7A 0.348 B-cell CLL/lymphoma 7A isoform b BCL7A 0.334 B-cell CLL/lymphoma 7A isoform a TNRC18 0.048 trinucleotide repeat containing 18 SETD1A 0.040 SET domain containing 1A HIVEP3 0.037 human immunodeficiency virus type I enhancer bindin... HIVEP3 0.037 human immunodeficiency virus type I enhancer bindin... MUC17 0.037 mucin 17 TCERG1 0.037 transcription elongation regulator 1 isoform 2 [Homo... TCERG1 0.037 transcription elongation regulator 1 isoform 1 [Homo... TOX 0.035 thymus high mobility group box protein TOX ACRC 0.035 ACRC protein DSPP 0.032 dentin sialophosphoprotein preproprotein LIG1 0.032 DNA ligase I TRDN 0.032 triadin C1orf113 0.029 SH3 domain-containing protein C1orf113 isoform 1 [H... C1orf113 0.029 SH3 domain-containing protein C1orf113 isoform 2 [H... BRD4 0.029 bromodomain-containing protein 4 isoform long NCOR2 0.027 nuclear receptor co-repressor 2 isoform 2 TCHHL1 0.027 trichohyalin-like 1 HOXA3 0.027 homeobox A3 isoform a HOXA3 0.027 homeobox A3 isoform a BASP1 0.027 brain abundant, membrane attached signal protein 1 [... MLLT3 0.027 myeloid/lymphoid or mixed-lineage leukemia (trithor... NT5C1B 0.027 5' nucleotidase, cytosolic IB isoform 2 EIF4G1 0.027 eukaryotic translation initiation factor 4 gamma, 1 ... EIF4G1 0.027 eukaryotic translation initiation factor 4 gamma, 1 ... EIF4G1 0.027 eukaryotic translation initiation factor 4 gamma, 1 ... EIF4G1 0.027 eukaryotic translation initiation factor 4 gamma, 1 ...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
