
| Name: KRTAP4-6 | Sequence: fasta or formatted (288aa) | NCBI GI: 169211815 | |
|
Description: PREDICTED: keratin associated protein 4.6
|
Referenced in:
| ||
|
Composition:
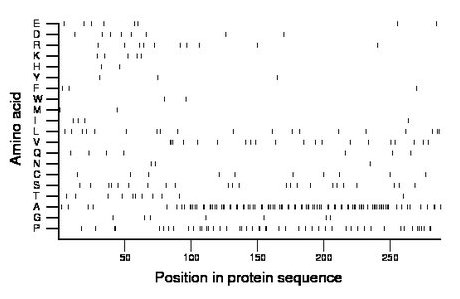
Amino acid Percentage Count Longest homopolymer A alanine 32.3 93 4 C cysteine 3.5 10 1 D aspartate 2.8 8 1 E glutamate 2.8 8 1 F phenylalanine 1.0 3 1 G glycine 2.4 7 1 H histidine 0.7 2 1 I isoleucine 1.4 4 1 K lysine 1.7 5 1 L leucine 6.6 19 2 M methionine 0.7 2 1 N asparagine 1.0 3 1 P proline 16.7 48 2 Q glutamine 2.4 7 1 R arginine 3.5 10 1 S serine 8.0 23 1 T threonine 3.8 11 1 V valine 6.9 20 3 W tryptophan 0.7 2 1 Y tyrosine 1.0 3 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: keratin associated protein 4.6 KRTAP4-9 0.458 PREDICTED: keratin associated protein 4-9 AMOT 0.103 angiomotin isoform 2 AMOT 0.103 angiomotin isoform 1 TAF4 0.100 TBP-associated factor 4 C4orf40 0.096 hypothetical protein LOC401137 KIAA0754 0.094 hypothetical protein LOC643314 SRCAP 0.090 Snf2-related CBP activator protein BAT2D1 0.089 HBxAg transactivated protein 2 SAMD1 0.089 sterile alpha motif domain containing 1 FLJ22184 0.087 PREDICTED: hypothetical protein LOC80164 BCORL1 0.087 BCL6 co-repressor-like 1 FLJ22184 0.085 PREDICTED: hypothetical protein FLJ22184 FLJ22184 0.083 PREDICTED: hypothetical protein FLJ22184 TMPRSS13 0.083 transmembrane protease, serine 13 YLPM1 0.077 YLP motif containing 1 FAM48B1 0.076 hypothetical protein LOC100130302 NACA 0.076 nascent polypeptide-associated complex alpha subuni... HOXA13 0.074 homeobox A13 LOC100291479 0.072 PREDICTED: hypothetical protein XP_002348186 FOXD1 0.068 forkhead box D1 CDKN1C 0.068 cyclin-dependent kinase inhibitor 1C isoform b [Hom... CDKN1C 0.068 cyclin-dependent kinase inhibitor 1C isoform b [Hom... CDKN1C 0.068 cyclin-dependent kinase inhibitor 1C isoform a MAZ 0.068 MYC-associated zinc finger protein isoform 2 MAZ 0.068 MYC-associated zinc finger protein isoform 1 COL6A3 0.061 alpha 3 type VI collagen isoform 5 precursor COL6A3 0.061 alpha 3 type VI collagen isoform 1 precursor COL6A3 0.061 alpha 3 type VI collagen isoform 4 precursor LOC100292122 0.059 PREDICTED: hypothetical protein XP_002345138Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
