
| Name: LOC730883 | Sequence: fasta or formatted (95aa) | NCBI GI: 169201702 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
Other entries for this name:
alt mRNA {95aa} PREDICTED: hypothetical protein | |||
|
Composition:
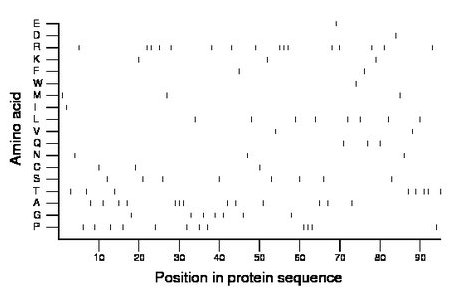
Amino acid Percentage Count Longest homopolymer A alanine 13.7 13 3 C cysteine 3.2 3 1 D aspartate 1.1 1 1 E glutamate 1.1 1 1 F phenylalanine 2.1 2 1 G glycine 7.4 7 1 H histidine 0.0 0 0 I isoleucine 1.1 1 1 K lysine 3.2 3 1 L leucine 8.4 8 1 M methionine 3.2 3 1 N asparagine 3.2 3 1 P proline 12.6 12 3 Q glutamine 3.2 3 1 R arginine 16.8 16 3 S serine 8.4 8 1 T threonine 8.4 8 2 V valine 2.1 2 1 W tryptophan 1.1 1 1 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC730883 1.000 PREDICTED: hypothetical protein FOXD2 0.053 forkhead box D2 MEFV 0.053 Mediterranean fever protein HCN2 0.047 hyperpolarization activated cyclic nucleotide-gated... COL10A1 0.047 type X collagen alpha 1 precursor DCAF10 0.047 WD repeat domain 32 LOC100291634 0.047 PREDICTED: hypothetical protein XP_002346062 LOC100291096 0.047 PREDICTED: hypothetical protein XP_002346952 LOC100287232 0.047 PREDICTED: hypothetical protein XP_002342816 LOC100287232 0.047 PREDICTED: hypothetical protein LOC728317 0.041 PREDICTED: hypothetical protein LOC728317 0.041 PREDICTED: hypothetical protein LOC728317 0.041 PREDICTED: hypothetical protein LOC728307 0.041 PREDICTED: hypothetical protein SORCS2 0.041 VPS10 domain receptor protein SORCS 2 MLL 0.041 myeloid/lymphoid or mixed-lineage leukemia protein [... FOXE1 0.041 forkhead box E1 COL25A1 0.041 collagen, type XXV, alpha 1 isoform 1 COL25A1 0.041 collagen, type XXV, alpha 1 isoform 2 C9orf69 0.041 hypothetical protein LOC90120 DLX2 0.035 distal-less homeobox 2 NKX1-1 0.035 PREDICTED: NK1 homeobox 1 HS3ST6 0.035 heparan sulfate (glucosamine) 3-O-sulfotransferase ... LOC100293961 0.035 PREDICTED: hypothetical protein LOC100290753 0.035 PREDICTED: hypothetical protein XP_002346471 LOC100289293 0.035 PREDICTED: hypothetical protein XP_002342302 ZDHHC1 0.035 zinc finger, DHHC-type containing 1 LOC100127891 0.029 PREDICTED: hypothetical protein LOC100131091 0.029 PREDICTED: hypothetical proteinHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
