
| Name: LOC100290753 | Sequence: fasta or formatted (138aa) | NCBI GI: 239747900 | |
|
Description: PREDICTED: hypothetical protein XP_002346471
| Not currently referenced in the text | ||
|
Composition:
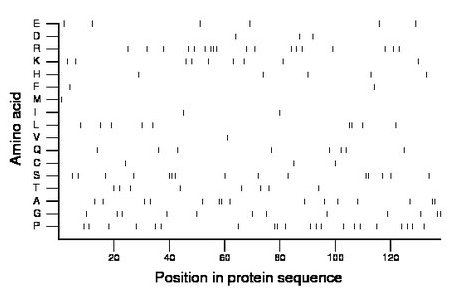
Amino acid Percentage Count Longest homopolymer A alanine 10.9 15 2 C cysteine 2.2 3 1 D aspartate 2.2 3 1 E glutamate 4.3 6 1 F phenylalanine 1.4 2 1 G glycine 8.7 12 2 H histidine 3.6 5 1 I isoleucine 1.4 2 1 K lysine 6.5 9 1 L leucine 6.5 9 2 M methionine 0.7 1 1 N asparagine 0.0 0 0 P proline 15.2 21 2 Q glutamine 5.8 8 1 R arginine 13.0 18 3 S serine 10.9 15 3 T threonine 5.8 8 1 V valine 0.7 1 1 W tryptophan 0.0 0 0 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002346471 LOC100293961 1.000 PREDICTED: hypothetical protein LOC100289293 1.000 PREDICTED: hypothetical protein XP_002342302 LOC100288017 0.347 PREDICTED: hypothetical protein XP_002343778 LOC100128382 0.179 PREDICTED: hypothetical protein LOC100128382 0.179 PREDICTED: hypothetical protein LOC100128382 0.179 PREDICTED: hypothetical protein GLTSCR1 0.073 glioma tumor suppressor candidate region gene 1 [Ho... PHLDA1 0.057 pleckstrin homology-like domain, family A, member 1 ... PCLO 0.057 piccolo isoform 2 PCLO 0.057 piccolo isoform 1 MAPK8IP3 0.053 mitogen-activated protein kinase 8 interacting prote... MAPK8IP3 0.053 mitogen-activated protein kinase 8 interacting prote... LOC643355 0.050 PREDICTED: hypothetical protein LOC643355 0.050 PREDICTED: hypothetical protein DDN 0.046 dendrin SRCAP 0.046 Snf2-related CBP activator protein LOC100292727 0.046 PREDICTED: hypothetical protein LOC100287920 0.046 PREDICTED: hypothetical protein XP_002344198 FLJ37078 0.046 hypothetical protein LOC222183 LOC400236 0.042 PREDICTED: hypothetical protein LOC400236 0.042 PREDICTED: hypothetical protein SRRM1 0.042 serine/arginine repetitive matrix 1 LOC100293385 0.038 PREDICTED: hypothetical protein LOC100290381 0.038 PREDICTED: hypothetical protein XP_002346884 LOC100289601 0.038 PREDICTED: hypothetical protein XP_002342725 CTDP1 0.038 CTD (carboxy-terminal domain, RNA polymerase II, pol... LOC100131414 0.038 PREDICTED: hypothetical protein LOC100131414 0.038 PREDICTED: hypothetical protein LOC100131414 0.038 PREDICTED: hypothetical proteinHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
