
| Name: LOC730456 | Sequence: fasta or formatted (174aa) | NCBI GI: 169166217 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
Other entries for this name:
alt prot {175aa} PREDICTED: hypothetical protein alt prot {197aa} PREDICTED: hypothetical protein | |||
|
Composition:
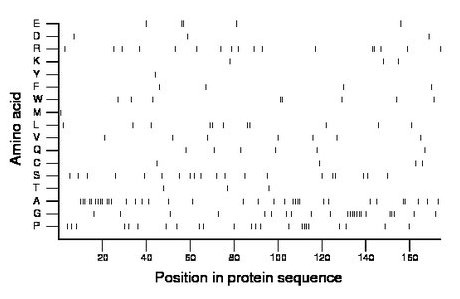
Amino acid Percentage Count Longest homopolymer A alanine 20.1 35 4 C cysteine 2.3 4 1 D aspartate 1.7 3 1 E glutamate 2.9 5 2 F phenylalanine 2.3 4 1 G glycine 13.2 23 7 H histidine 0.0 0 0 I isoleucine 0.0 0 0 K lysine 1.7 3 1 L leucine 6.3 11 2 M methionine 0.6 1 1 N asparagine 0.0 0 0 P proline 13.2 23 4 Q glutamine 3.4 6 1 R arginine 9.8 17 2 S serine 11.5 20 2 T threonine 1.7 3 1 V valine 4.0 7 1 W tryptophan 4.6 8 2 Y tyrosine 0.6 1 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC730456 0.619 PREDICTED: hypothetical protein LOC730456 0.619 PREDICTED: hypothetical protein VASP 0.056 vasodilator-stimulated phosphoprotein LOC100289763 0.053 PREDICTED: hypothetical protein XP_002347867 LOC100289681 0.053 PREDICTED: hypothetical protein XP_002343623 LOC100292172 0.053 PREDICTED: hypothetical protein LOC100127913 0.050 PREDICTED: hypothetical protein FLJ31945 0.050 PREDICTED: hypothetical protein LOC100127913 0.050 PREDICTED: hypothetical protein FLJ31945 0.050 PREDICTED: hypothetical protein LOC100127913 0.050 PREDICTED: hypothetical protein CASKIN2 0.047 cask-interacting protein 2 isoform b CASKIN2 0.047 cask-interacting protein 2 isoform a FLJ22184 0.044 PREDICTED: hypothetical protein FLJ22184 FLJ22184 0.044 PREDICTED: hypothetical protein FLJ22184 FLJ22184 0.044 PREDICTED: hypothetical protein LOC80164 ATXN2 0.044 ataxin 2 LOC338758 0.044 PREDICTED: hypothetical protein LOC338758 0.044 PREDICTED: hypothetical protein LOC100291533 0.044 PREDICTED: hypothetical protein XP_002347080 LOC338758 0.044 PREDICTED: hypothetical protein LOC100288002 0.044 PREDICTED: hypothetical protein XP_002342946 FOXE1 0.044 forkhead box E1 FAM48B1 0.044 hypothetical protein LOC100130302 MYO15A 0.041 myosin XV ZNF703 0.041 zinc finger protein 703 FLJ31945 0.041 PREDICTED: hypothetical protein MARK4 0.041 MAP/microtubule affinity-regulating kinase 4 EOMES 0.041 eomesoderminHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
