
| Name: LOC730456 | Sequence: fasta or formatted (197aa) | NCBI GI: 169167080 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
Other entries for this name:
alt prot {175aa} PREDICTED: hypothetical protein alt prot {174aa} PREDICTED: hypothetical protein | |||
|
Composition:
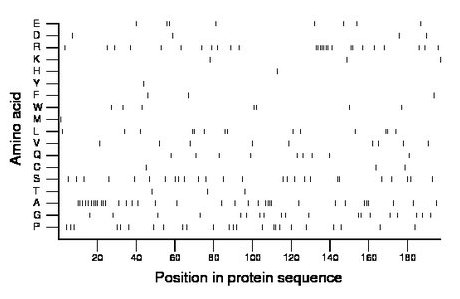
Amino acid Percentage Count Longest homopolymer A alanine 17.8 35 4 C cysteine 1.5 3 1 D aspartate 2.0 4 1 E glutamate 4.1 8 2 F phenylalanine 1.5 3 1 G glycine 9.6 19 2 H histidine 0.5 1 1 I isoleucine 0.0 0 0 K lysine 1.5 3 1 L leucine 7.1 14 2 M methionine 0.5 1 1 N asparagine 0.0 0 0 P proline 12.2 24 2 Q glutamine 4.6 9 1 R arginine 13.7 27 7 S serine 13.2 26 2 T threonine 1.5 3 1 V valine 4.6 9 1 W tryptophan 3.6 7 2 Y tyrosine 0.5 1 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC730456 0.653 PREDICTED: hypothetical protein LOC730456 0.560 PREDICTED: hypothetical protein FAM48B1 0.048 hypothetical protein LOC100130302 DDN 0.045 dendrin LOC100289763 0.042 PREDICTED: hypothetical protein XP_002347867 LOC100289681 0.042 PREDICTED: hypothetical protein XP_002343623 LOC100292172 0.042 PREDICTED: hypothetical protein LOC100291337 0.040 PREDICTED: hypothetical protein XP_002348329 LOC100287998 0.040 PREDICTED: hypothetical protein XP_002342281 ANKRD33B 0.040 PREDICTED: ankyrin repeat domain 33B MICALL1 0.037 molecule interacting with Rab13 MYO15A 0.037 myosin XV LOC100128979 0.037 PREDICTED: hypothetical protein LOC100128979 0.037 PREDICTED: hypothetical protein DACT3 0.037 thymus expressed gene 3-like ANKRD56 0.037 ankyrin repeat domain 56 LOC441086 0.034 PREDICTED: hypothetical protein AGAP2 0.032 centaurin, gamma 1 isoform PIKE-L LOC100292157 0.032 PREDICTED: hypothetical protein LOC100129571 0.032 PREDICTED: similar to hCG1646049 LOC100290926 0.032 PREDICTED: hypothetical protein XP_002347568 LOC100129571 0.032 PREDICTED: similar to hCG1646049 LOC100288243 0.032 PREDICTED: hypothetical protein LOC100288995 0.032 PREDICTED: hypothetical protein XP_002343379 LOC100129571 0.032 PREDICTED: similar to hCG1646049 LOC100288243 0.032 PREDICTED: hypothetical protein XP_002342233 FOXE1 0.032 forkhead box E1 ADAM8 0.032 ADAM metallopeptidase domain 8 precursor C17orf65 0.032 hypothetical protein LOC339201Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
