
| Name: LOC730456 | Sequence: fasta or formatted (175aa) | NCBI GI: 113416229 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
Other entries for this name:
alt prot {174aa} PREDICTED: hypothetical protein alt prot {197aa} PREDICTED: hypothetical protein | |||
|
Composition:
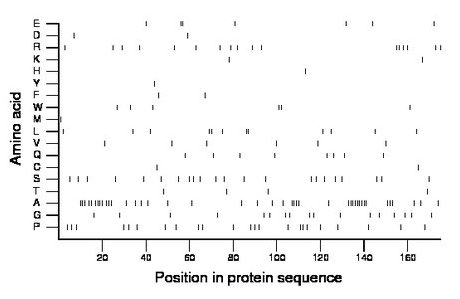
Amino acid Percentage Count Longest homopolymer A alanine 24.0 42 9 C cysteine 1.1 2 1 D aspartate 1.1 2 1 E glutamate 4.0 7 2 F phenylalanine 1.1 2 1 G glycine 9.7 17 1 H histidine 0.6 1 1 I isoleucine 0.0 0 0 K lysine 1.1 2 1 L leucine 6.9 12 2 M methionine 0.6 1 1 N asparagine 0.0 0 0 P proline 13.1 23 2 Q glutamine 4.6 8 1 R arginine 9.7 17 2 S serine 12.6 22 1 T threonine 2.3 4 1 V valine 3.4 6 1 W tryptophan 3.4 6 2 Y tyrosine 0.6 1 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC730456 0.748 PREDICTED: hypothetical protein LOC730456 0.641 PREDICTED: hypothetical protein FAM48B1 0.070 hypothetical protein LOC100130302 DDN 0.067 dendrin FOXE1 0.064 forkhead box E1 LOC100290274 0.061 PREDICTED: hypothetical protein XP_002346984 LOC100288645 0.061 PREDICTED: hypothetical protein XP_002342846 FOXL2 0.061 forkhead box L2 FOXD1 0.058 forkhead box D1 LOC100289681 0.052 PREDICTED: hypothetical protein XP_002343623 LOC100292157 0.049 PREDICTED: hypothetical protein LOC100289763 0.049 PREDICTED: hypothetical protein XP_002347867 LOC100290926 0.049 PREDICTED: hypothetical protein XP_002347568 LOC100288995 0.049 PREDICTED: hypothetical protein XP_002343379 LOC100292172 0.049 PREDICTED: hypothetical protein C17orf96 0.049 hypothetical protein LOC100170841 LOC100294444 0.046 PREDICTED: similar to family with sequence similari... FIGNL2 0.046 fidgetin-like 2 SALL3 0.046 sal-like 3 FAM48B2 0.046 family with sequence similarity 48, member B2 [Homo... MYO15A 0.046 myosin XV VASP 0.046 vasodilator-stimulated phosphoprotein LOC100129098 0.046 PREDICTED: hypothetical protein BHLHE41 0.046 basic helix-loop-helix domain containing, class B, 3... TAF4 0.046 TBP-associated factor 4 C2orf55 0.043 hypothetical protein LOC343990 CDC2L5 0.043 cell division cycle 2-like 5 isoform 1 CDC2L5 0.043 cell division cycle 2-like 5 isoform 2 DACT3 0.043 thymus expressed gene 3-likeHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
