
| Name: LOC729199 | Sequence: fasta or formatted (261aa) | NCBI GI: 169160983 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
|
Composition:
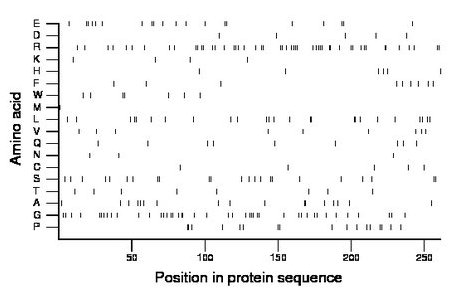
Amino acid Percentage Count Longest homopolymer A alanine 6.5 17 2 C cysteine 1.9 5 1 D aspartate 1.9 5 1 E glutamate 7.7 20 2 F phenylalanine 3.4 9 1 G glycine 16.5 43 3 H histidine 2.3 6 1 I isoleucine 0.0 0 0 K lysine 1.5 4 1 L leucine 9.6 25 2 M methionine 0.4 1 1 N asparagine 1.1 3 1 P proline 6.5 17 2 Q glutamine 3.4 9 1 R arginine 19.2 50 3 S serine 8.8 23 2 T threonine 3.1 8 1 V valine 3.4 9 1 W tryptophan 2.7 7 2 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein SFRS16 0.037 splicing factor, arginine/serine-rich 16 LOC339192 0.035 PREDICTED: hypothetical protein LOC100292370 0.033 PREDICTED: hypothetical protein LOC100291835 0.033 PREDICTED: hypothetical protein XP_002346266 LOC339192 0.031 PREDICTED: hypothetical protein LOC339192 0.031 PREDICTED: hypothetical protein TMEM88B 0.029 PREDICTED: hypothetical protein LOC643965 TMEM88B 0.029 PREDICTED: hypothetical protein LOC643965 HRNR 0.027 hornerin MBD2 0.027 methyl-CpG binding domain protein 2 testis-specific i... MBD2 0.027 methyl-CpG binding domain protein 2 isoform 1 LOC100292292 0.027 PREDICTED: hypothetical protein LOC100291384 0.027 PREDICTED: hypothetical protein XP_002347156 LOC100291533 0.027 PREDICTED: hypothetical protein XP_002347080 LOC100287578 0.027 PREDICTED: hypothetical protein XP_002343004 LOC100288002 0.027 PREDICTED: hypothetical protein XP_002342946 CLIC6 0.027 chloride intracellular channel 6 SRRM1 0.027 serine/arginine repetitive matrix 1 LOC728046 0.025 PREDICTED: hypothetical protein UBAP2L 0.025 ubiquitin associated protein 2-like isoform a [Homo... UBAP2L 0.025 ubiquitin associated protein 2-like isoform b [Homo... SRRM2 0.025 splicing coactivator subunit SRm300 MLLT4 0.023 myeloid/lymphoid or mixed-lineage leukemia (trithora... LOC100291843 0.023 PREDICTED: hypothetical protein XP_002344913 LOC390595 0.023 PREDICTED: similar to ubiquitin-associated protein ... LOC390595 0.023 PREDICTED: similar to ubiquitin-associated protein ... LOC390595 0.023 PREDICTED: hypothetical protein SMAD6 0.023 SMAD family member 6 isoform 1 ZNF497 0.023 zinc finger protein 497Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
