
| Name: C1orf110 | Sequence: fasta or formatted (302aa) | NCBI GI: 148806888 | |
|
Description: hypothetical protein LOC339512
| Not currently referenced in the text | ||
|
Composition:
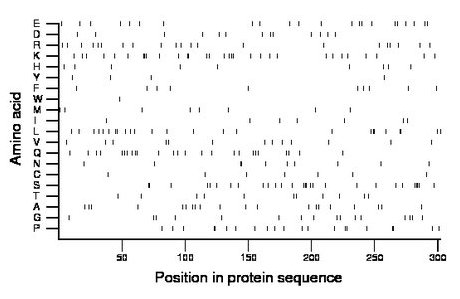
Amino acid Percentage Count Longest homopolymer A alanine 6.6 20 1 C cysteine 2.3 7 1 D aspartate 4.3 13 1 E glutamate 7.3 22 1 F phenylalanine 3.3 10 1 G glycine 5.3 16 1 H histidine 3.0 9 1 I isoleucine 2.3 7 1 K lysine 10.3 31 3 L leucine 8.3 25 2 M methionine 2.6 8 1 N asparagine 3.3 10 2 P proline 7.0 21 2 Q glutamine 8.3 25 2 R arginine 7.0 21 1 S serine 9.3 28 4 T threonine 3.6 11 1 V valine 4.3 13 1 W tryptophan 0.3 1 1 Y tyrosine 1.3 4 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 hypothetical protein LOC339512 GPATCH2 0.027 G patch domain containing 2 MAP4 0.019 microtubule-associated protein 4 isoform 5 MAP4 0.019 microtubule-associated protein 4 isoform 4 MAP4 0.019 microtubule-associated protein 4 isoform 2 MAP4 0.019 microtubule-associated protein 4 isoform 1 MAP7D3 0.017 MAP7 domain containing 3 OGFRL1 0.017 opioid growth factor receptor-like 1 RSL1D1 0.015 ribosomal L1 domain containing 1 IWS1 0.015 IWS1 homolog SERPINH1 0.014 serine (or cysteine) proteinase inhibitor, clade H, ... NEFH 0.014 neurofilament, heavy polypeptide 200kDa MINK1 0.014 misshapen/NIK-related kinase isoform 4 KIAA1826 0.014 hypothetical protein LOC84437 ZZEF1 0.014 zinc finger, ZZ type with EF hand domain 1 RBM28 0.014 RNA binding motif protein 28 ABCF1 0.012 ATP-binding cassette, sub-family F, member 1 isoform... ABCF1 0.012 ATP-binding cassette, sub-family F, member 1 isoform... FKBPL 0.012 WAF-1/CIP1 stabilizing protein 39 VCPIP1 0.012 valosin containing protein (p97)/p47 complex interac... TTC28 0.012 tetratricopeptide repeat domain 28 LZTS2 0.012 leucine zipper, putative tumor suppressor 2 HAUS6 0.012 HAUS augmin-like complex, subunit 6 MDN1 0.012 MDN1, midasin homolog ANKRD35 0.010 ankyrin repeat domain 35 RBM17 0.010 RNA binding motif protein 17 RBM17 0.010 RNA binding motif protein 17 PDE4B 0.010 phosphodiesterase 4B isoform 1 PDE4B 0.010 phosphodiesterase 4B isoform 3 PDE4B 0.010 phosphodiesterase 4B isoform 2Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
