
| Name: LOC642669 | Sequence: fasta or formatted (130aa) | NCBI GI: 89037235 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
|
Composition:
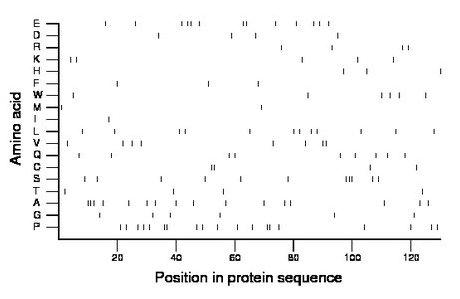
Amino acid Percentage Count Longest homopolymer A alanine 12.3 16 3 C cysteine 3.1 4 2 D aspartate 3.1 4 1 E glutamate 10.0 13 2 F phenylalanine 2.3 3 1 G glycine 4.6 6 1 H histidine 2.3 3 1 I isoleucine 0.8 1 1 K lysine 3.8 5 1 L leucine 9.2 12 1 M methionine 1.5 2 1 N asparagine 0.0 0 0 P proline 14.6 19 2 Q glutamine 6.9 9 1 R arginine 3.1 4 1 S serine 8.5 11 3 T threonine 3.1 4 1 V valine 6.2 8 2 W tryptophan 4.6 6 1 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC100289194 0.992 PREDICTED: hypothetical protein XP_002343934 LOC644525 0.992 PREDICTED: hypothetical protein LOC644525 0.925 PREDICTED: hypothetical protein C2orf27A 0.894 hypothetical protein LOC29798 C2orf27B 0.823 hypothetical protein LOC408029 RGL4 0.138 ral guanine nucleotide dissociation stimulator-like ... CDRT15 0.079 CMT1A duplicated region transcript 15 RALGDS 0.063 ral guanine nucleotide dissociation stimulator isofo... RALGDS 0.063 ral guanine nucleotide dissociation stimulator isof... ARHGAP29 0.047 PTPL1-associated RhoGAP 1 FSCB 0.039 fibrous sheath CABYR binding protein LOC100287920 0.039 PREDICTED: hypothetical protein XP_002344198 ARAP1 0.039 ArfGAP with RhoGAP domain, ankyrin repeat and PH dom... LOC100294050 0.039 PREDICTED: hypothetical protein DGKK 0.039 diacylglycerol kinase kappa NACA 0.035 nascent polypeptide-associated complex alpha subuni... TAF4 0.031 TBP-associated factor 4 SAMD1 0.031 sterile alpha motif domain containing 1 SPATA2L 0.031 spermatogenesis associated 2-like SPEN 0.031 spen homolog, transcriptional regulator CHRM4 0.028 cholinergic receptor, muscarinic 4 VCX 0.028 variable charge, X chromosome VCX2 0.028 variable charge, X-linked 2 VCX3A 0.028 variable charge, X-linked 3A C6orf132 0.028 PREDICTED: chromosome 6 open reading frame 132 [Hom... FLJ22184 0.028 PREDICTED: hypothetical protein FLJ22184 C6orf132 0.028 PREDICTED: chromosome 6 open reading frame 132 [Hom... C6orf132 0.028 PREDICTED: hypothetical protein LOC647024 SHANK1 0.028 SH3 and multiple ankyrin repeat domains 1Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
