
| Name: C2orf27B | Sequence: fasta or formatted (209aa) | NCBI GI: 47551356 | |
|
Description: hypothetical protein LOC408029
| Not currently referenced in the text | ||
|
Composition:
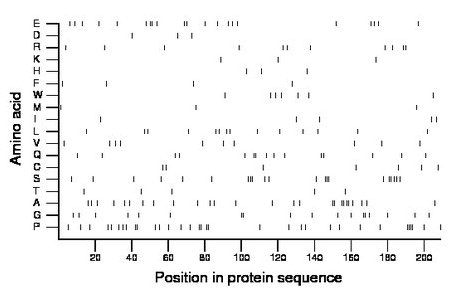
Amino acid Percentage Count Longest homopolymer A alanine 12.4 26 2 C cysteine 3.3 7 1 D aspartate 1.4 3 1 E glutamate 10.0 21 2 F phenylalanine 1.9 4 1 G glycine 8.6 18 2 H histidine 1.4 3 1 I isoleucine 2.4 5 1 K lysine 1.4 3 1 L leucine 6.7 14 1 M methionine 1.4 3 1 N asparagine 0.0 0 0 P proline 15.8 33 4 Q glutamine 7.2 15 2 R arginine 5.3 11 2 S serine 10.0 21 3 T threonine 2.4 5 1 V valine 4.8 10 1 W tryptophan 3.3 7 1 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 hypothetical protein LOC408029 C2orf27A 0.926 hypothetical protein LOC29798 LOC644525 0.837 PREDICTED: hypothetical protein LOC100289194 0.507 PREDICTED: hypothetical protein XP_002343934 LOC644525 0.507 PREDICTED: hypothetical protein LOC642669 0.500 PREDICTED: hypothetical protein RGL4 0.091 ral guanine nucleotide dissociation stimulator-like ... CDRT15 0.062 CMT1A duplicated region transcript 15 TAF4 0.053 TBP-associated factor 4 LOC100129129 0.048 PREDICTED: hypothetical protein RALGDS 0.045 ral guanine nucleotide dissociation stimulator isofo... RALGDS 0.045 ral guanine nucleotide dissociation stimulator isof... NANOS3 0.041 nanos homolog 3 AATK 0.033 apoptosis-associated tyrosine kinase PTPN23 0.033 protein tyrosine phosphatase, non-receptor type 23 [... DGKK 0.033 diacylglycerol kinase kappa ARHGAP29 0.033 PTPL1-associated RhoGAP 1 FSCB 0.031 fibrous sheath CABYR binding protein KCNN3 0.029 small conductance calcium-activated potassium chann... SAMD1 0.029 sterile alpha motif domain containing 1 FLJ22184 0.029 PREDICTED: hypothetical protein FLJ22184 ROBO3 0.029 roundabout, axon guidance receptor, homolog 3 POU6F2 0.029 POU domain, class 6, transcription factor 2 FOXL1 0.029 forkhead box L1 C22orf26 0.029 hypothetical protein LOC55267 ADAMTSL4 0.026 thrombospondin repeat containing 1 isoform 1 ADAMTSL4 0.026 thrombospondin repeat containing 1 isoform 2 NAP5 0.026 Nck-associated protein 5 isoform 1 FMNL1 0.026 formin-like 1 FHDC1 0.024 FH2 domain containing 1Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
