
| Name: CDRT15 | Sequence: fasta or formatted (188aa) | NCBI GI: 56090618 | |
|
Description: CMT1A duplicated region transcript 15
| Not currently referenced in the text | ||
|
Composition:
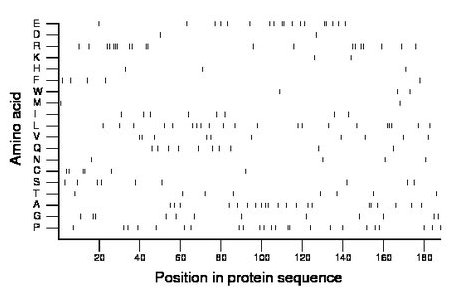
Amino acid Percentage Count Longest homopolymer A alanine 11.2 21 2 C cysteine 3.2 6 2 D aspartate 1.1 2 1 E glutamate 9.6 18 2 F phenylalanine 2.7 5 1 G glycine 6.4 12 2 H histidine 1.6 3 1 I isoleucine 4.3 8 1 K lysine 1.1 2 1 L leucine 11.2 21 3 M methionine 1.1 2 1 N asparagine 2.1 4 1 P proline 12.8 24 2 Q glutamine 5.3 10 1 R arginine 10.6 20 3 S serine 4.8 9 1 T threonine 4.3 8 1 V valine 5.3 10 2 W tryptophan 1.6 3 1 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 CMT1A duplicated region transcript 15 LOC644525 0.188 PREDICTED: hypothetical protein C2orf27B 0.072 hypothetical protein LOC408029 LOC100289194 0.064 PREDICTED: hypothetical protein XP_002343934 LOC644525 0.064 PREDICTED: hypothetical protein C2orf27A 0.064 hypothetical protein LOC29798 LOC642669 0.055 PREDICTED: hypothetical protein DGKK 0.047 diacylglycerol kinase kappa BASP1 0.039 brain abundant, membrane attached signal protein 1 [... RALGDS 0.039 ral guanine nucleotide dissociation stimulator isofo... RALGDS 0.039 ral guanine nucleotide dissociation stimulator isof... LOC342346 0.030 hypothetical protein LOC342346 SH3BP1 0.028 SH3-domain binding protein 1 EIF4G1 0.028 eukaryotic translation initiation factor 4 gamma, 1 ... EIF4G1 0.028 eukaryotic translation initiation factor 4 gamma, 1 ... EIF4G1 0.028 eukaryotic translation initiation factor 4 gamma, 1 ... EIF4G1 0.028 eukaryotic translation initiation factor 4 gamma, 1 ... EIF4G1 0.028 eukaryotic translation initiation factor 4 gamma, 1 ... RIN3 0.028 Ras and Rab interactor 3 FLJ39639 0.025 PREDICTED: hypothetical protein FLJ39639 FLJ39639 0.025 PREDICTED: hypothetical protein FLJ39639 FLJ39639 0.025 PREDICTED: hypothetical protein LOC283876 RSPH4A 0.025 radial spoke head 4 homolog A isoform 2 RSPH4A 0.025 radial spoke head 4 homolog A isoform 1 LOC401934 0.022 PREDICTED: hypothetical protein LOC401934 0.022 PREDICTED: hypothetical protein RGL4 0.022 ral guanine nucleotide dissociation stimulator-like ... SPAG1 0.022 sperm associated antigen 1 SPAG1 0.022 sperm associated antigen 1 TTN 0.022 titin isoform N2-AHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
