
| Name: VCX2 | Sequence: fasta or formatted (139aa) | NCBI GI: 49619239 | |
|
Description: variable charge, X-linked 2
|
Referenced in:
| ||
|
Composition:
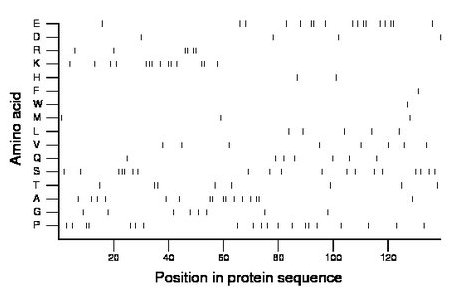
Amino acid Percentage Count Longest homopolymer A alanine 11.5 16 2 C cysteine 0.0 0 0 D aspartate 2.9 4 1 E glutamate 12.2 17 2 F phenylalanine 0.7 1 1 G glycine 5.8 8 1 H histidine 1.4 2 1 I isoleucine 0.0 0 0 K lysine 10.1 14 3 L leucine 3.6 5 1 M methionine 2.2 3 1 N asparagine 0.0 0 0 P proline 14.4 20 2 Q glutamine 5.0 7 1 R arginine 4.3 6 2 S serine 13.7 19 3 T threonine 5.8 8 2 V valine 5.8 8 1 W tryptophan 0.7 1 1 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 variable charge, X-linked 2 VCX 0.881 variable charge, X chromosome VCX3A 0.881 variable charge, X-linked 3A VCX3B 0.874 variable charge, X-linked 3B VCY1B 0.759 variable charge, Y-linked 1B VCY 0.759 variable charge, Y-linked AEBP1 0.079 adipocyte enhancer binding protein 1 precursor [Homo... NEFH 0.071 neurofilament, heavy polypeptide 200kDa LOC727838 0.063 PREDICTED: similar to cancer/testis antigen family ... COLEC12 0.063 collectin sub-family member 12 MAP7D1 0.063 MAP7 domain containing 1 GIGYF2 0.059 GRB10 interacting GYF protein 2 isoform a KIAA1602 0.059 hypothetical protein LOC57701 POLD3 0.059 DNA-directed DNA polymerase delta 3 CASKIN2 0.055 cask-interacting protein 2 isoform b CASKIN2 0.055 cask-interacting protein 2 isoform a COL19A1 0.055 alpha 1 type XIX collagen precursor AAK1 0.055 AP2 associated kinase 1 DKK3 0.051 dickkopf homolog 3 precursor DKK3 0.051 dickkopf homolog 3 precursor DKK3 0.051 dickkopf homolog 3 precursor LOC727838 0.051 PREDICTED: similar to cancer/testis antigen family ... COL3A1 0.051 collagen type III alpha 1 preproprotein CCNK 0.051 cyclin K isoform 1 BAT2D1 0.051 HBxAg transactivated protein 2 AP3D1 0.047 adaptor-related protein complex 3, delta 1 subunit ... EPB41L2 0.047 erythrocyte membrane protein band 4.1-like 2 isoform ... TCOF1 0.047 Treacher Collins-Franceschetti syndrome 1 isoform f... TCOF1 0.047 Treacher Collins-Franceschetti syndrome 1 isoform e... TCOF1 0.047 Treacher Collins-Franceschetti syndrome 1 isoform d...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
