
| Name: GTPBP1 | Sequence: fasta or formatted (669aa) | NCBI GI: 82546879 | |
|
Description: GTP binding protein 1
|
Referenced in: Additional Guanine Nucleotide-binding Proteins
| ||
|
Composition:
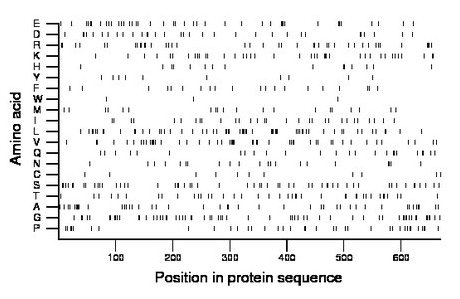
Amino acid Percentage Count Longest homopolymer A alanine 6.9 46 6 C cysteine 1.6 11 1 D aspartate 5.1 34 2 E glutamate 6.1 41 3 F phenylalanine 3.0 20 1 G glycine 9.4 63 2 H histidine 2.2 15 2 I isoleucine 4.5 30 1 K lysine 6.0 40 2 L leucine 9.1 61 3 M methionine 3.1 21 1 N asparagine 3.1 21 2 P proline 5.7 38 3 Q glutamine 3.7 25 2 R arginine 6.1 41 3 S serine 8.5 57 2 T threonine 6.3 42 2 V valine 7.5 50 3 W tryptophan 0.4 3 1 Y tyrosine 1.5 10 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 GTP binding protein 1 GTPBP2 0.368 GTP binding protein 2 TUFM 0.051 Tu translation elongation factor, mitochondrial prec... HBS1L 0.044 Hsp70 subfamily B suppressor 1-like protein isoform... HBS1L 0.044 Hsp70 subfamily B suppressor 1-like protein isoform 1... EEF1A2 0.043 eukaryotic translation elongation factor 1 alpha 2 [H... EEF1A1 0.041 eukaryotic translation elongation factor 1 alpha 1 [H... GSPT1 0.038 G1 to S phase transition 1 isoform 3 GSPT1 0.038 G1 to S phase transition 1 isoform 2 GSPT1 0.038 G1 to S phase transition 1 isoform 1 GSPT2 0.033 peptide chain release factor 3 LOC100293909 0.031 PREDICTED: similar to eukaryotic translation elonga... EEFSEC 0.026 eukaryotic elongation factor, selenocysteine-tRNA-sp... EIF5B 0.019 eukaryotic translation initiation factor 5B SNIP 0.011 SNAP25-interacting protein SFPQ 0.009 splicing factor proline/glutamine rich (polypyrimidin... LOC100288426 0.008 PREDICTED: hypothetical protein XP_002343430 LOC100132369 0.008 PREDICTED: hypothetical protein LOC100132369 0.008 PREDICTED: hypothetical protein LOC100132369 0.008 PREDICTED: hypothetical protein MAP4 0.008 microtubule-associated protein 4 isoform 5 MAP4 0.008 microtubule-associated protein 4 isoform 2 MAP4 0.008 microtubule-associated protein 4 isoform 1 CCDC88B 0.008 coiled-coil domain containing 88 KCNQ1 0.007 potassium voltage-gated channel, KQT-like subfamily,... KDM6B 0.007 lysine (K)-specific demethylase 6B PAWR 0.007 PRKC, apoptosis, WT1, regulator SLC38A10 0.007 solute carrier family 38, member 10 isoform b LOC100292454 0.006 PREDICTED: similar to hCG2019076 FUSSEL18 0.006 PREDICTED: functional smad suppressing element 18 [...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
