
| Name: TFPT | Sequence: fasta or formatted (253aa) | NCBI GI: 7019371 | |
|
Description: TCF3 (E2A) fusion partner
| Not currently referenced in the text | ||
|
Composition:
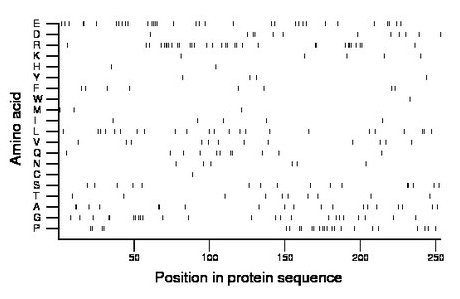
Amino acid Percentage Count Longest homopolymer A alanine 7.5 19 2 C cysteine 0.4 1 1 D aspartate 4.7 12 2 E glutamate 13.4 34 4 F phenylalanine 3.2 8 1 G glycine 9.5 24 2 H histidine 0.8 2 1 I isoleucine 2.0 5 1 K lysine 2.0 5 1 L leucine 9.1 23 2 M methionine 1.2 3 1 N asparagine 2.4 6 1 P proline 9.5 24 2 Q glutamine 4.3 11 2 R arginine 13.0 33 4 S serine 6.3 16 2 T threonine 4.0 10 1 V valine 4.7 12 1 W tryptophan 0.4 1 1 Y tyrosine 1.6 4 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 TCF3 (E2A) fusion partner SPTBN4 0.033 spectrin, beta, non-erythrocytic 4 isoform sigma1 [... SNRNP70 0.031 U1 small nuclear ribonucleoprotein 70 kDa SHANK3 0.031 SH3 and multiple ankyrin repeat domains 3 ZC3H18 0.029 zinc finger CCCH-type containing 18 FMNL1 0.029 formin-like 1 HCN2 0.029 hyperpolarization activated cyclic nucleotide-gated... DDIT3 0.027 DNA-damage-inducible transcript 3 EIF3A 0.027 eukaryotic translation initiation factor 3, subunit 1... SETBP1 0.027 SET binding protein 1 isoform a CCDC85B 0.027 hepatitis delta antigen-interacting protein A SMG5 0.027 SMG5 homolog nonsense mediated mRNA decay factor [H... PSPC1 0.027 paraspeckle protein 1 DAAM1 0.027 dishevelled-associated activator of morphogenesis 1 ... NEUROG1 0.025 neurogenin 1 ZC3H13 0.025 zinc finger CCCH-type containing 13 MAP4K4 0.025 mitogen-activated protein kinase kinase kinase kinas... INO80E 0.025 INO80 complex subunit E OGFR 0.025 opioid growth factor receptor LOC100286959 0.025 PREDICTED: hypothetical protein XP_002343921 LOC100128074 0.025 PREDICTED: hypothetical protein UPF3B 0.025 UPF3 regulator of nonsense transcripts homolog B iso... RBM25 0.023 RNA binding motif protein 25 FAM71E2 0.023 hypothetical protein LOC284418 TUSC1 0.023 tumor suppressor candidate 1 FLJ37078 0.023 hypothetical protein LOC222183 PHLDB3 0.023 pleckstrin homology-like domain, family B, member 3... ARHGEF18 0.023 Rho/Rac guanine nucleotide exchange factor 18 isofo... ARHGEF18 0.023 Rho/Rac guanine nucleotide exchange factor 18 isofor... CACNA1G 0.023 voltage-dependent calcium channel alpha 1G subunit i...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
