
| Name: IGSF3 | Sequence: fasta or formatted (1214aa) | NCBI GI: 55953133 | |
|
Description: immunoglobulin superfamily, member 3 isoform 1
|
Referenced in: Additional Genes in Development
| ||
Other entries for this name:
alt prot [1194aa] immunoglobulin superfamily, member 3 isoform 2 | |||
|
Composition:
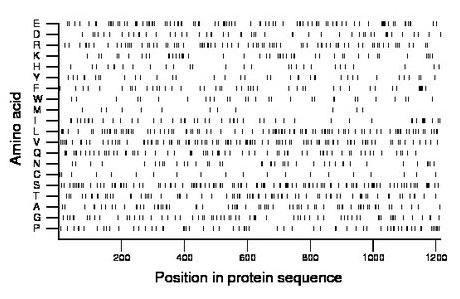
Amino acid Percentage Count Longest homopolymer A alanine 5.5 67 2 C cysteine 1.6 20 1 D aspartate 4.4 53 7 E glutamate 8.2 99 6 F phenylalanine 3.8 46 2 G glycine 5.5 67 2 H histidine 2.4 29 2 I isoleucine 4.2 51 2 K lysine 3.8 46 1 L leucine 8.3 101 2 M methionine 1.1 13 1 N asparagine 3.4 41 2 P proline 5.2 63 2 Q glutamine 5.5 67 1 R arginine 6.8 82 2 S serine 10.0 121 3 T threonine 6.3 76 2 V valine 8.6 105 2 W tryptophan 2.4 29 1 Y tyrosine 3.1 38 2 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 immunoglobulin superfamily, member 3 isoform 1 [Homo... IGSF3 0.977 immunoglobulin superfamily, member 3 isoform 2 [Homo... LOC100292114 0.267 PREDICTED: similar to immunoglobulin superfamily, m... IGSF2 0.244 immunoglobulin superfamily, member 2 IGSF8 0.105 immunoglobulin superfamily, member 8 PTGFRN 0.090 prostaglandin F2 receptor negative regulator LOC100132705 0.088 PREDICTED: similar to immunoglobulin superfamily, m... LOC100132900 0.088 PREDICTED: similar to KIAA0466 protein HSPG2 0.015 heparan sulfate proteoglycan 2 C10orf72 0.009 hypothetical protein LOC196740 isoform 2 C10orf72 0.009 hypothetical protein LOC196740 isoform 1 CSRNP3 0.008 cysteine-serine-rich nuclear protein 3 PIGR 0.008 polymeric immunoglobulin receptor precursor TTN 0.008 titin isoform N2-A LOC100290415 0.007 PREDICTED: hypothetical protein XP_002347493 MXRA8 0.007 matrix-remodelling associated 8 ZNF526 0.007 zinc finger protein 526 TTBK1 0.007 tau tubulin kinase 1 SETD1B 0.007 SET domain containing 1B VPRBP 0.006 HIV-1 Vpr binding protein CENPB 0.006 centromere protein B HOMEZ 0.006 homeodomain leucine zipper protein PRDM2 0.006 retinoblastoma protein-binding zinc finger protein i... PRDM2 0.006 retinoblastoma protein-binding zinc finger protein i... PRDM2 0.006 retinoblastoma protein-binding zinc finger protein i... ARID4B 0.006 AT rich interactive domain 4B isoform 1 OBSL1 0.006 obscurin-like 1 LOC100291786 0.006 PREDICTED: similar to hCG2043206 LOC440871 0.006 PREDICTED: similar to hCG2043206 BTNL2 0.006 butyrophilin-like 2Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
