
| Name: LOC100132900 | Sequence: fasta or formatted (155aa) | NCBI GI: 239746626 | |
|
Description: PREDICTED: similar to KIAA0466 protein
|
Referenced in: Additional Genes in Development
| ||
|
Composition:
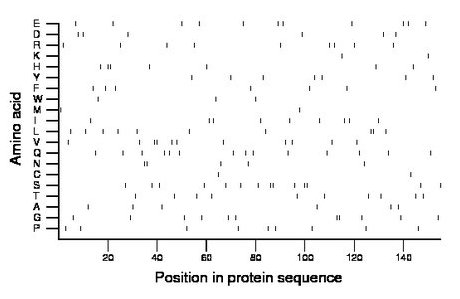
Amino acid Percentage Count Longest homopolymer A alanine 3.9 6 1 C cysteine 1.3 2 1 D aspartate 3.9 6 1 E glutamate 7.1 11 1 F phenylalanine 4.5 7 1 G glycine 7.1 11 2 H histidine 4.5 7 2 I isoleucine 5.8 9 1 K lysine 1.3 2 1 L leucine 6.5 10 2 M methionine 1.3 2 1 N asparagine 3.2 5 2 P proline 5.8 9 1 Q glutamine 9.0 14 1 R arginine 5.8 9 1 S serine 9.0 14 2 T threonine 6.5 10 1 V valine 7.1 11 2 W tryptophan 1.9 3 1 Y tyrosine 4.5 7 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: similar to KIAA0466 protein LOC100132705 0.973 PREDICTED: similar to immunoglobulin superfamily, m... IGSF3 0.717 immunoglobulin superfamily, member 3 isoform 1 [Homo... IGSF3 0.717 immunoglobulin superfamily, member 3 isoform 2 [Homo... IGSF2 0.493 immunoglobulin superfamily, member 2 IGSF8 0.363 immunoglobulin superfamily, member 8 PTGFRN 0.207 prostaglandin F2 receptor negative regulator LOC100290415 0.043 PREDICTED: hypothetical protein XP_002347493 LOC100292114 0.030 PREDICTED: similar to immunoglobulin superfamily, m... LOC100130811 0.027 PREDICTED: similar to hCG2038921 LOC100130811 0.027 PREDICTED: similar to hCG2038921 HMCN1 0.027 hemicentin 1 NCAM1 0.023 neural cell adhesion molecule 1 isoform 1 NCAM1 0.023 neural cell adhesion molecule 1 isoform 2 NCAM1 0.023 neural cell adhesion molecule 1 isoform 3 TTN 0.020 titin isoform N2-A NPTN 0.020 neuroplastin isoform d precursor NPTN 0.020 neuroplastin isoform c precursor NPTN 0.020 neuroplastin isoform a precursor NPTN 0.020 neuroplastin isoform b precursor SCN4B 0.020 sodium channel, voltage-gated, type IV, beta isoform... VSIG1 0.020 V-set and immunoglobulin domain containing 1 OBSL1 0.017 obscurin-like 1 CNTN1 0.017 contactin 1 isoform 1 precursor CNTN1 0.017 contactin 1 isoform 2 precursor CH25H 0.017 cholesterol 25-hydroxylase CD274 0.013 CD274 molecule TTN 0.013 titin isoform novex-2 TTN 0.013 titin isoform novex-1 TTN 0.013 titin isoform N2-BHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
