
| Name: SLPI | Sequence: fasta or formatted (132aa) | NCBI GI: 4507065 | |
|
Description: secretory leukocyte peptidase inhibitor precursor
|
Referenced in:
| ||
|
Composition:
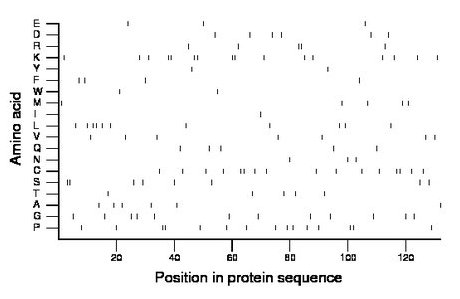
Amino acid Percentage Count Longest homopolymer A alanine 4.5 6 1 C cysteine 12.1 16 2 D aspartate 4.5 6 1 E glutamate 2.3 3 1 F phenylalanine 3.0 4 1 G glycine 9.1 12 1 H histidine 0.0 0 0 I isoleucine 0.8 1 1 K lysine 12.1 16 2 L leucine 8.3 11 2 M methionine 3.8 5 1 N asparagine 2.3 3 1 P proline 11.4 15 2 Q glutamine 3.8 5 1 R arginine 3.8 5 2 S serine 6.1 8 2 T threonine 3.8 5 1 V valine 5.3 7 1 W tryptophan 1.5 2 1 Y tyrosine 1.5 2 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 secretory leukocyte peptidase inhibitor precursor [Ho... WFDC5 0.176 WAP four-disulfide core domain 5 precursor WFDC3 0.154 WAP four-disulfide core domain 3 precursor PI3 0.132 elafin preproprotein WFDC2 0.132 WAP four-disulfide core domain 2 precursor WFDC12 0.117 WAP four-disulfide core domain 12 precursor WFDC8 0.088 WAP four-disulfide core domain 8 precursor WFDC8 0.088 WAP four-disulfide core domain 8 precursor SPINLW1 0.066 serine peptidase inhibitor-like, with Kunitz and WAP... SPINLW1 0.059 serine peptidase inhibitor-like, with Kunitz and WAP... WFDC1 0.051 WAP four-disulfide core domain 1 precursor WFDC6 0.048 WAP four-disulfide core domain 6 precursor WFIKKN2 0.044 WFIKKN2 protein KAL1 0.044 Kallmann syndrome 1 protein precursor NPNT 0.037 nephronectin WFIKKN1 0.033 WAP, follistatin/kazal, immunoglobulin, kunitz and n... UMODL1 0.033 uromodulin-like 1 isoform 1 precursor UMODL1 0.033 uromodulin-like 1 isoform 2 precursor PRG4 0.029 proteoglycan 4 isoform C PRG4 0.029 proteoglycan 4 isoform A CRIM1 0.029 cysteine-rich motor neuron 1 ERBB3 0.029 erbB-3 isoform 1 precursor DLL3 0.029 delta-like 3 protein isoform 1 precursor DLL3 0.029 delta-like 3 protein isoform 2 precursor LOC728318 0.022 PREDICTED: hypothetical protein LOC728318 0.022 PREDICTED: hypothetical protein LOC100127930 0.022 PREDICTED: hypothetical protein LOC728318 0.022 PREDICTED: hypothetical protein LOC100127930 0.022 PREDICTED: hypothetical protein SORL1 0.022 sortilin-related receptor containing LDLR class A rep...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
