
| Name: WFDC12 | Sequence: fasta or formatted (111aa) | NCBI GI: 20069858 | |
|
Description: WAP four-disulfide core domain 12 precursor
|
Referenced in:
| ||
|
Composition:
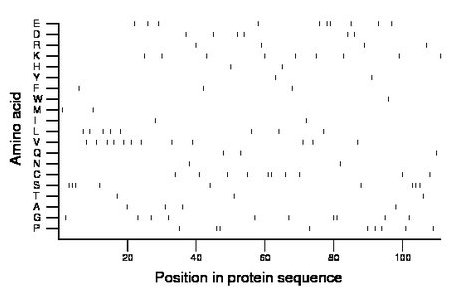
Amino acid Percentage Count Longest homopolymer A alanine 3.6 4 1 C cysteine 9.0 10 2 D aspartate 5.4 6 1 E glutamate 9.0 10 2 F phenylalanine 2.7 3 1 G glycine 9.0 10 2 H histidine 1.8 2 1 I isoleucine 1.8 2 1 K lysine 8.1 9 1 L leucine 7.2 8 1 M methionine 1.8 2 1 N asparagine 1.8 2 1 P proline 8.1 9 2 Q glutamine 2.7 3 1 R arginine 3.6 4 1 S serine 8.1 9 3 T threonine 2.7 3 1 V valine 10.8 12 1 W tryptophan 0.9 1 1 Y tyrosine 1.8 2 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 WAP four-disulfide core domain 12 precursor SLPI 0.148 secretory leukocyte peptidase inhibitor precursor [Ho... WFDC5 0.125 WAP four-disulfide core domain 5 precursor WFDC8 0.116 WAP four-disulfide core domain 8 precursor WFDC8 0.116 WAP four-disulfide core domain 8 precursor PI3 0.102 elafin preproprotein SPINLW1 0.088 serine peptidase inhibitor-like, with Kunitz and WAP... WFDC3 0.083 WAP four-disulfide core domain 3 precursor WFDC2 0.083 WAP four-disulfide core domain 2 precursor WFDC6 0.074 WAP four-disulfide core domain 6 precursor KAL1 0.056 Kallmann syndrome 1 protein precursor SPINLW1 0.051 serine peptidase inhibitor-like, with Kunitz and WAP... WFIKKN2 0.051 WFIKKN2 protein WFIKKN1 0.037 WAP, follistatin/kazal, immunoglobulin, kunitz and n... WFDC1 0.032 WAP four-disulfide core domain 1 precursor UMODL1 0.032 uromodulin-like 1 isoform 1 precursor UMODL1 0.032 uromodulin-like 1 isoform 2 precursor SPP1 0.028 secreted phosphoprotein 1 isoform b SPP1 0.028 secreted phosphoprotein 1 isoform a C1orf113 0.014 SH3 domain-containing protein C1orf113 isoform 1 [H... C1orf113 0.014 SH3 domain-containing protein C1orf113 isoform 2 [H... SFRS14 0.014 splicing factor, arginine/serine-rich 14 SFRS14 0.014 splicing factor, arginine/serine-rich 14 ZFP64 0.014 zinc finger protein 64 isoform d LTBP3 0.014 latent transforming growth factor beta binding prot... LTBP3 0.014 latent transforming growth factor beta binding prote... FBLN5 0.009 fibulin 5 precursor ZZEF1 0.009 zinc finger, ZZ type with EF hand domain 1 CASZ1 0.009 castor homolog 1, zinc finger isoform a ENPP2 0.009 autotaxin isoform 3 preproproteinHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
