
| Name: LOC100127930 | Sequence: fasta or formatted (143aa) | NCBI GI: 169210045 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
Other entries for this name:
alt prot {447aa} PREDICTED: hypothetical protein alt mRNA {143aa} PREDICTED: hypothetical protein | |||
|
Composition:
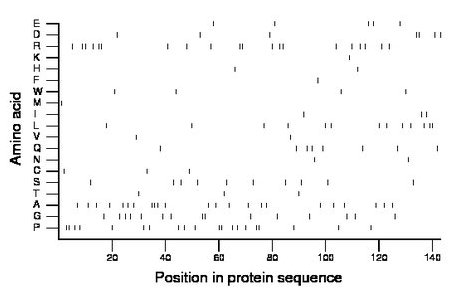
Amino acid Percentage Count Longest homopolymer A alanine 16.1 23 3 C cysteine 2.1 3 1 D aspartate 4.9 7 2 E glutamate 3.5 5 1 F phenylalanine 0.7 1 1 G glycine 10.5 15 2 H histidine 1.4 2 1 I isoleucine 2.1 3 1 K lysine 0.7 1 1 L leucine 9.1 13 2 M methionine 0.7 1 1 N asparagine 1.4 2 1 P proline 14.0 20 2 Q glutamine 5.6 8 1 R arginine 14.0 20 2 S serine 7.0 10 1 T threonine 2.1 3 1 V valine 1.4 2 1 W tryptophan 2.8 4 1 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC100127930 1.000 PREDICTED: hypothetical protein LOC100127930 0.830 PREDICTED: hypothetical protein LOC100290023 0.076 PREDICTED: hypothetical protein XP_002348147 LOC100290265 0.054 PREDICTED: hypothetical protein XP_002346787 TAF4 0.054 TBP-associated factor 4 MAPK7 0.054 mitogen-activated protein kinase 7 isoform 1 MAPK7 0.054 mitogen-activated protein kinase 7 isoform 1 MAPK7 0.054 mitogen-activated protein kinase 7 isoform 1 MAPK7 0.054 mitogen-activated protein kinase 7 isoform 2 SASH1 0.051 SAM and SH3 domain containing 1 DMWD 0.051 dystrophia myotonica-containing WD repeat motif prot... TTMA 0.051 hypothetical protein LOC645369 LOC100293818 0.051 PREDICTED: hypothetical protein SAMD1 0.051 sterile alpha motif domain containing 1 LOC100292447 0.047 PREDICTED: similar to coiled-coil domain containing... LOC100131102 0.047 PREDICTED: hypothetical protein ZFPM1 0.047 zinc finger protein, multitype 1 LOC100128440 0.043 PREDICTED: hypothetical protein LOC100128440 0.043 PREDICTED: hypothetical protein LOC100128440 0.043 PREDICTED: hypothetical protein LOC646396 0.043 PREDICTED: similar to hCG2042704 MLL2 0.043 myeloid/lymphoid or mixed-lineage leukemia 2 MAST3 0.043 microtubule associated serine/threonine kinase 3 [H... LOC100131102 0.043 PREDICTED: hypothetical protein LOC100130360 0.043 PREDICTED: hypothetical protein SRCAP 0.043 Snf2-related CBP activator protein ZIC5 0.043 zinc finger protein of the cerebellum 5 DDN 0.043 dendrin LOC284297 0.040 hypothetical protein LOC284297Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
