
| Name: WFDC6 | Sequence: fasta or formatted (86aa) | NCBI GI: 22779936 | |
|
Description: WAP four-disulfide core domain 6 precursor
|
Referenced in:
| ||
|
Composition:
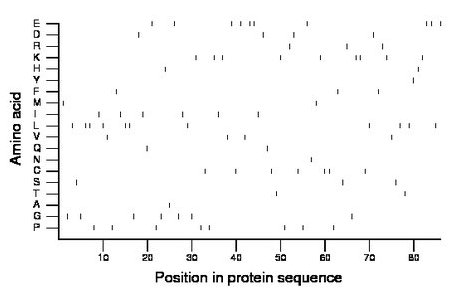
Amino acid Percentage Count Longest homopolymer A alanine 1.2 1 1 C cysteine 8.1 7 2 D aspartate 4.7 4 1 E glutamate 11.6 10 2 F phenylalanine 3.5 3 1 G glycine 8.1 7 1 H histidine 2.3 2 1 I isoleucine 7.0 6 1 K lysine 10.5 9 2 L leucine 12.8 11 2 M methionine 2.3 2 1 N asparagine 1.2 1 1 P proline 9.3 8 1 Q glutamine 2.3 2 1 R arginine 3.5 3 1 S serine 3.5 3 1 T threonine 2.3 2 1 V valine 4.7 4 1 W tryptophan 0.0 0 0 Y tyrosine 1.2 1 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 WAP four-disulfide core domain 6 precursor SPINLW1 0.404 serine peptidase inhibitor-like, with Kunitz and WAP... SPINLW1 0.242 serine peptidase inhibitor-like, with Kunitz and WAP... WFDC8 0.118 WAP four-disulfide core domain 8 precursor WFDC8 0.118 WAP four-disulfide core domain 8 precursor WFDC12 0.099 WAP four-disulfide core domain 12 precursor SLPI 0.081 secretory leukocyte peptidase inhibitor precursor [Ho... PI3 0.050 elafin preproprotein WFDC5 0.043 WAP four-disulfide core domain 5 precursor WFDC3 0.043 WAP four-disulfide core domain 3 precursor UMODL1 0.037 uromodulin-like 1 isoform 1 precursor UMODL1 0.037 uromodulin-like 1 isoform 2 precursor SAP130 0.037 Sin3A-associated protein, 130kDa isoform b SAP130 0.037 Sin3A-associated protein, 130kDa isoform a EMR1 0.031 egf-like module containing, mucin-like, hormone rece... LOC100293983 0.025 PREDICTED: similar to mucin, partial WFDC2 0.019 WAP four-disulfide core domain 2 precursor SLC12A9 0.019 solute carrier family 12 (potassium/chloride transpo... SERPINC1 0.012 serpin peptidase inhibitor, clade C, member 1 R3HDM1 0.012 R3H domain containing 1 WFDC10A 0.012 WAP four-disulfide core domain 10A precursor LTBP3 0.006 latent transforming growth factor beta binding prot... LTBP3 0.006 latent transforming growth factor beta binding prote... GRM8 0.006 glutamate receptor, metabotropic 8 isoform b precur... GRM8 0.006 glutamate receptor, metabotropic 8 isoform a precur... PRAMEF7 0.006 PRAME family member 7 LOC729516 0.006 PREDICTED: similar to Putative PRAME family member ... PRAMEF8 0.006 PRAME family member 8Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
