
| Name: LOC100293844 | Sequence: fasta or formatted (105aa) | NCBI GI: 239756961 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
|
Composition:
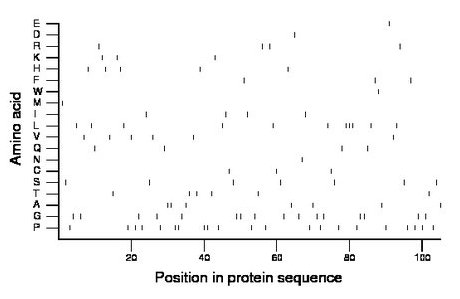
Amino acid Percentage Count Longest homopolymer A alanine 6.7 7 2 C cysteine 2.9 3 1 D aspartate 1.0 1 1 E glutamate 1.0 1 1 F phenylalanine 2.9 3 1 G glycine 15.2 16 2 H histidine 4.8 5 1 I isoleucine 3.8 4 1 K lysine 2.9 3 1 L leucine 10.5 11 3 M methionine 1.0 1 1 N asparagine 1.0 1 1 P proline 20.0 21 2 Q glutamine 3.8 4 1 R arginine 3.8 4 1 S serine 6.7 7 1 T threonine 5.7 6 1 V valine 5.7 6 1 W tryptophan 1.0 1 1 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC100287030 0.917 PREDICTED: hypothetical protein XP_002343628 LOC100291048 0.663 PREDICTED: hypothetical protein XP_002347845 LOC100292352 0.332 PREDICTED: hypothetical protein LOC100287760 0.332 PREDICTED: hypothetical protein LOC100287760 0.332 PREDICTED: hypothetical protein XP_002343356 WBP11 0.068 WW domain binding protein 11 ATN1 0.054 atrophin-1 ATN1 0.054 atrophin-1 DIAPH1 0.054 diaphanous 1 isoform 2 DIAPH1 0.054 diaphanous 1 isoform 1 FBXL19 0.039 F-box and leucine-rich repeat protein 19 CCNK 0.039 cyclin K isoform 1 TPRX1 0.039 tetra-peptide repeat homeobox LOC728888 0.039 PREDICTED: similar to acyl-CoA synthetase medium-ch... LOC643355 0.039 PREDICTED: hypothetical protein DAZAP2 0.034 DAZ associated protein 2 isoform d ZC3H4 0.034 zinc finger CCCH-type containing 4 C9orf75 0.034 hypothetical protein LOC286262 isoform 2 C9orf75 0.034 hypothetical protein LOC286262 isoform 1 KIAA1522 0.034 hypothetical protein LOC57648 PPP1R10 0.034 protein phosphatase 1, regulatory subunit 10 PRR13 0.034 proline rich 13 isoform 1 TAF4 0.029 TBP-associated factor 4 LOC100131102 0.029 PREDICTED: hypothetical protein LOC100131102 0.029 PREDICTED: hypothetical protein LOC100131102 0.029 PREDICTED: hypothetical protein TRIM28 0.029 tripartite motif-containing 28 protein DAZAP2 0.029 DAZ associated protein 2 isoform a NPAS1 0.029 neuronal PAS domain protein 1Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
