
| Name: FAM153B | Sequence: fasta or formatted (387aa) | NCBI GI: 118918431 | |
|
Description: hypothetical protein LOC202134
| Not currently referenced in the text | ||
|
Composition:
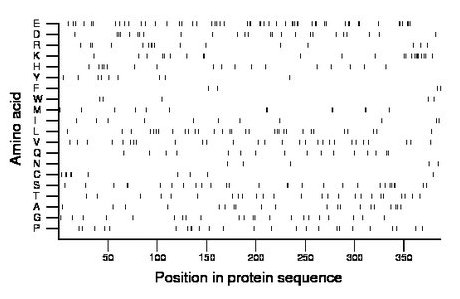
Amino acid Percentage Count Longest homopolymer A alanine 5.2 20 1 C cysteine 2.6 10 2 D aspartate 6.5 25 1 E glutamate 14.0 54 3 F phenylalanine 1.0 4 1 G glycine 5.7 22 1 H histidine 4.4 17 1 I isoleucine 2.8 11 1 K lysine 5.9 23 3 L leucine 8.0 31 2 M methionine 3.4 13 2 N asparagine 1.3 5 1 P proline 6.5 25 2 Q glutamine 4.1 16 2 R arginine 4.1 16 2 S serine 7.2 28 2 T threonine 6.5 25 1 V valine 7.2 28 1 W tryptophan 1.3 5 1 Y tyrosine 2.3 9 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 hypothetical protein LOC202134 FAM153A 0.757 hypothetical protein LOC285596 FAM153C 0.149 hypothetical protein LOC653316 RP1L1 0.025 retinitis pigmentosa 1-like 1 FSCB 0.022 fibrous sheath CABYR binding protein LOC400986 0.019 PREDICTED: protein immuno-reactive with anti-PTH po... ANKRD36B 0.019 ankyrin repeat domain 36B OPTN 0.017 optineurin OPTN 0.017 optineurin OPTN 0.017 optineurin OPTN 0.017 optineurin MDC1 0.017 mediator of DNA-damage checkpoint 1 MICAL3 0.017 microtubule associated monoxygenase, calponin and L... LOC400986 0.016 PREDICTED: protein immuno-reactive with anti-PTH po... LOC100133923 0.013 PREDICTED: similar to UPF0634 protein C SLTM 0.013 modulator of estrogen induced transcription isoform ... MAGED2 0.013 melanoma antigen family D, 2 MAGED2 0.013 melanoma antigen family D, 2 MAGED2 0.013 melanoma antigen family D, 2 AKAP12 0.013 A kinase (PRKA) anchor protein 12 isoform 1 AKAP12 0.013 A kinase (PRKA) anchor protein 12 isoform 2 GNL2 0.013 guanine nucleotide binding protein-like 2 (nucleolar)... MYH10 0.013 myosin, heavy polypeptide 10, non-muscle RBM33 0.013 RNA binding motif protein 33 SCARF1 0.012 scavenger receptor class F, member 1 isoform 1 precu... SCARF1 0.012 scavenger receptor class F, member 1 isoform 5 prec... PPIG 0.012 peptidylprolyl isomerase G DZIP3 0.012 DAZ interacting protein 3, zinc finger WBP4 0.012 WW domain-containing binding protein 4 BOD1L 0.010 biorientation of chromosomes in cell division 1-like...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
