
| Name: LOC100294366 | Sequence: fasta or formatted (79aa) | NCBI GI: 239752105 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
|
Composition:
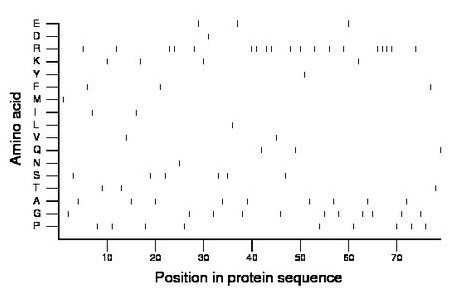
Amino acid Percentage Count Longest homopolymer A alanine 11.4 9 1 C cysteine 0.0 0 0 D aspartate 1.3 1 1 E glutamate 3.8 3 1 F phenylalanine 3.8 3 1 G glycine 13.9 11 1 H histidine 0.0 0 0 I isoleucine 2.5 2 1 K lysine 5.1 4 1 L leucine 1.3 1 1 M methionine 1.3 1 1 N asparagine 1.3 1 1 P proline 11.4 9 1 Q glutamine 3.8 3 1 R arginine 24.1 19 4 S serine 7.6 6 1 T threonine 3.8 3 1 V valine 2.5 2 1 W tryptophan 0.0 0 0 Y tyrosine 1.3 1 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC100293809 1.000 PREDICTED: hypothetical protein LOC100288573 0.599 PREDICTED: hypothetical protein XP_002343799 DEDD2 0.080 death effector domain-containing DNA binding protei... LOC100291634 0.073 PREDICTED: hypothetical protein XP_002346062 LOC100291096 0.073 PREDICTED: hypothetical protein XP_002346952 LOC100287232 0.073 PREDICTED: hypothetical protein XP_002342816 LOC100287232 0.073 PREDICTED: hypothetical protein C19orf29 0.066 chromosome 19 open reading frame 29 C19orf29 0.066 chromosome 19 open reading frame 29 NPHS2 0.058 podocin LOC100292916 0.058 PREDICTED: hypothetical protein LOC100290781 0.058 PREDICTED: hypothetical protein XP_002347101 LOC100286938 0.058 PREDICTED: hypothetical protein XP_002342984 LOC728896 0.058 PREDICTED: hypothetical protein LOC100287002 0.058 PREDICTED: hypothetical protein XP_002343745 LOC100293776 0.051 PREDICTED: hypothetical protein LOC100288658 0.051 PREDICTED: hypothetical protein XP_002342750 LOC100288658 0.051 PREDICTED: hypothetical protein SMAD6 0.051 SMAD family member 6 isoform 1 BAG1 0.051 BCL2-associated athanogene isoform 1L BHLHA15 0.051 basic helix-loop-helix family, member a15 FAM38A 0.051 family with sequence similarity 38, member A KIAA0368 0.044 KIAA0368 protein RRS1 0.044 homolog of yeast ribosome biogenesis regulatory prot... LOC100292122 0.044 PREDICTED: hypothetical protein XP_002345138 LOC100286986 0.044 PREDICTED: hypothetical protein XP_002344191 LOC100286967 0.044 PREDICTED: hypothetical protein XP_002342247 ZNF318 0.044 zinc finger protein 318 LOC728392 0.036 hypothetical protein LOC728392Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
